Initial solution: See references above.
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
{
return(exp(a*log(x)));
}
{
double gamma = 1.4;
double R = 287.058;
double rho_ref = 1.1612055171196529;
double p_ref = 100000.0;
double grav_x = 0.0;
double grav_y = 9.8;
int HB = 0;
int NI,NJ,ndims;
char ip_file_type[50]; strcpy(ip_file_type,"ascii");
FILE *in;
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) {
printf("Error: Input file \"solver.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")) {
while (strcmp(word, "end")) {
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) fscanf(in,"%d",&ndims);
else if (!strcmp(word, "size")) {
fscanf(in,"%d",&NI);
fscanf(in,"%d",&NJ);
} else if (!strcmp(word, "ip_file_type")) fscanf(in,"%s",ip_file_type);
}
} else printf("Error: Illegal format in solver.inp. Crash and burn!\n");
}
fclose(in);
printf("Reading file \"physics.inp\"...\n");
in = fopen("physics.inp","r");
if (!in) {
printf("Error: Input file \"physics.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")) {
while (strcmp(word, "end")) {
fscanf(in,"%s",word);
if (!strcmp(word, "rho_ref")) fscanf(in,"%lf",&rho_ref);
else if (!strcmp(word, "p_ref" )) fscanf(in,"%lf",&p_ref );
else if (!strcmp(word, "gamma" )) fscanf(in,"%lf",&gamma );
else if (!strcmp(word, "R" )) fscanf(in,"%lf",&R );
else if (!strcmp(word, "HB" )) fscanf(in,"%d" ,&HB );
else if (!strcmp(word, "gravity")) {
fscanf(in,"%lf",&grav_x );
fscanf(in,"%lf",&grav_y );
}
}
} else printf("Error: Illegal format in physics.inp. Crash and burn!\n");
}
fclose(in);
if (ndims != 2) {
printf("ndims is not 2 in solver.inp. this code is to generate 2D initial conditions\n");
return(0);
}
if (HB != 2) {
printf("Error: Specify \"HB\" as 2 in physics.inp.\n");
}
printf("Grid:\t\t\t%d X %d\n",NI,NJ);
printf("Reference density and pressure: %lf, %lf.\n",rho_ref,p_ref);
int i,j;
double dx = 1000.0 / ((double)(NI-1));
double dy = 1000.0 / ((double)(NJ-1));
double *x, *y, *u0, *u1, *u2, *u3;
x = (double*) calloc (NI , sizeof(double));
y = (double*) calloc (NJ , sizeof(double));
u0 = (double*) calloc (NI*NJ, sizeof(double));
u1 = (double*) calloc (NI*NJ, sizeof(double));
u2 = (double*) calloc (NI*NJ, sizeof(double));
u3 = (double*) calloc (NI*NJ, sizeof(double));
double xc = 500;
double yc = 350;
double Cp = gamma * R / (gamma-1.0);
double tc = 0.5;
double pi = 4.0*atan(1.0);
double rc = 250.0;
double T_ref = p_ref / (R * rho_ref);
for (i = 0; i < NI; i++){
for (j = 0; j < NJ; j++){
x[i] = i*dx;
y[j] = j*dy;
int p = NJ*i + j;
double r = sqrt((x[i]-xc)*(x[i]-xc)+(y[j]-yc)*(y[j]-yc));
double dtheta = (r>rc ? 0.0 : (0.5*tc*(1.0+cos(pi*r/rc))) );
double theta = T_ref + dtheta;
double Pexner = 1.0 - (grav_y*y[j])/(Cp*T_ref);
double rho = (p_ref/(R*theta)) *
raiseto(Pexner, (1.0/(gamma-1.0)));
double E = rho * (R/(gamma-1.0)) * theta*Pexner;
u0[p] = rho;
u1[p] = 0.0;
u2[p] = 0.0;
u3[p] = E;
}
}
FILE *out;
if (!strcmp(ip_file_type,"ascii")) {
printf("Writing ASCII initial solution file initial.inp\n");
out = fopen("initial.inp","w");
for (i = 0; i < NI; i++) fprintf(out,"%lf ",x[i]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) fprintf(out,"%lf ",y[j]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u0[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u1[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u2[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u3[p]);
}
}
fprintf(out,"\n");
fclose(out);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary initial solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x,sizeof(double),NI,out);
fwrite(y,sizeof(double),NJ,out);
double *U = (double*) calloc (4*NI*NJ,sizeof(double));
for (i=0; i < NI; i++) {
for (j = 0; j < NJ; j++) {
int p = NJ*i + j;
int q = NI*j + i;
U[4*q+0] = u0[p];
U[4*q+1] = u1[p];
U[4*q+2] = u2[p];
U[4*q+3] = u3[p];
}
}
fwrite(U,sizeof(double),4*NI*NJ,out);
free(U);
fclose(out);
}
free(x);
free(y);
free(u0);
free(u1);
free(u2);
free(u3);
return(0);
}
#include <stdio.h>
#include <stdlib.h>
#include <string.h>
#include <math.h>
typedef struct _parameters_{
double grav_x, grav_y, R, gamma, P_ref, rho_ref;
int HB;
} Parameters;
void IncrementFilename(char *f)
{
if (f[10] == '9') {
f[10] = '0';
if (f[9] == '9') {
f[9] = '0';
if (f[8] == '9') {
f[8] = '0';
if (f[7] == '9') {
f[7] = '0';
if (f[6] == '9') {
f[6] = '0';
fprintf(stderr,"Warning: file increment hit max limit. Resetting to zero.\n");
} else {
f[6]++;
}
} else {
f[7]++;
}
} else {
f[8]++;
}
} else {
f[9]++;
}
} else {
f[10]++;
}
}
{
return(exp(a*log(x)));
}
void WriteTecplot2D(
int nvars,
int imax,
int jmax,
double *x,
double *u,
char *f)
{
printf("\tWriting tecplot solution file %s.\n",f);
FILE *out;
out = fopen(f,"w");
if (!out) {
fprintf(stderr,"Error: could not open %s for writing.\n",f);
return;
}
double *X = x;
double *Y = x+imax;
fprintf(out,"VARIABLES=\"I\",\"J\",\"X\",\"Y\",");
fprintf(out,"\"RHO\",\"U\",\"V\",\"P\",");
fprintf(out,"\"THETA\",\"RHO0\",\"P0\",");
fprintf(out,"\"PI0\",\"THETA0\",\n");
fprintf(out,"ZONE I=%d,J=%d,F=POINT\n",imax,jmax);
int i,j;
for (j=0; j<jmax; j++) {
for (i=0; i<imax; i++) {
int v, p = i + imax*j;
fprintf(out,"%4d %4d ",i,j);
fprintf(out,"%1.16E %1.16E ",X[i],Y[j]);
for (v=0; v<nvars; v++) fprintf(out,"%1.16E ",u[nvars*p+v]);
fprintf(out,"\n");
}
}
fclose(out);
return;
}
void WriteText2D(int nvars,int imax, int jmax,double *x,double *u,char *f)
{
printf("\tWriting text solution file %s.\n",f);
FILE *out;
out = fopen(f,"w");
if (!out) {
fprintf(stderr,"Error: could not open %s for writing.\n",f);
return;
}
double *X = x;
double *Y = x+imax;
int i,j;
for (j=0; j<jmax; j++) {
for (i=0; i<imax; i++) {
int v, p = i + imax*j;
fprintf(out,"%4d %4d ",i,j);
fprintf(out,"%1.16E %1.16E ",X[i],Y[j]);
for (v=0; v<nvars; v++) fprintf(out,"%1.16E ",u[nvars*p+v]);
fprintf(out,"\n");
}
}
fclose(out);
return;
}
int PostProcess(char *fname, char *oname, void *p, int flag)
{
Parameters *params = (Parameters*) p;
FILE *in; in = fopen(fname,"rb");
if (!in) return(-1);
printf("Reading file %s.\n",fname);
int ndims, nvars;
double *U,*x;
fread(&ndims,sizeof(int),1,in);
fread(&nvars,sizeof(int),1,in);
if (ndims != 2) {
printf("Error: ndims in %s not equal to 2!\n",fname);
return(1);
}
if (nvars != 4) {
printf("Error: nvars in %s not equal to 4!\n",fname);
return(1);
}
int dims[ndims];
fread(dims,sizeof(int),ndims,in);
printf("Dimensions: %d x %d\n",dims[0],dims[1]);
printf("Nvars : %d\n",nvars);
x = (double*) calloc (dims[0]+dims[1] ,sizeof(double));
U = (double*) calloc (dims[0]*dims[1]*nvars ,sizeof(double));
fread(x,sizeof(double),dims[0]+dims[1] ,in);
fread(U,sizeof(double),dims[0]*dims[1]*nvars,in);
fclose(in);
int imax = dims[0];
int jmax = dims[1];
int evars = 5;
double *Q = (double*) calloc ((nvars+evars)*imax*jmax,sizeof(double));
int i, j;
double *X = x;
double *Y = x+imax;
double grav_y = params->grav_y;
double R = params->R;
double gamma = params->gamma;
double P_ref = params->P_ref;
double rho_ref = params->rho_ref;
double T_ref = P_ref / (R*rho_ref);
double inv_gamma_m1 = 1.0 / (gamma-1.0);
double Cp = gamma * inv_gamma_m1 * R;
for (i=0; i<imax; i++) {
for (j=0; j<jmax; j++) {
int p = i + imax*j;
double rho0, theta0, Pexner, P0;
theta0 = T_ref;
Pexner = 1.0 - (grav_y*Y[j])/(Cp*T_ref);
rho0 = (P_ref/(R*theta0)) *
raiseto(Pexner, inv_gamma_m1);
P0 = P_ref *
raiseto(Pexner, gamma*inv_gamma_m1);
double rho, uvel, vvel, E, P, theta;
rho = U[nvars*p+0];
uvel = U[nvars*p+1] / rho;
vvel = U[nvars*p+2] / rho;
E = U[nvars*p+3];
P = (gamma-1.0) * (E - 0.5*rho*(uvel*uvel+vvel*vvel));
theta = (E-0.5*rho*(uvel*uvel+vvel*vvel))/(Pexner*rho) * ((gamma-1.0)/R);
Q[(nvars+evars)*p+0] = rho;
Q[(nvars+evars)*p+1] = uvel;
Q[(nvars+evars)*p+2] = vvel;
Q[(nvars+evars)*p+3] = P;
Q[(nvars+evars)*p+4] = theta;
Q[(nvars+evars)*p+5] = rho0;
Q[(nvars+evars)*p+6] = P0;
Q[(nvars+evars)*p+7] = Pexner;
Q[(nvars+evars)*p+8] = theta0;
}
}
else WriteText2D (nvars+evars,imax,jmax,x,Q,oname);
free(U);
free(Q);
free(x);
}
{
FILE *out1, *out2, *in, *inputs;
char filename[50], op_file_format[50], tecfile[50], overwrite[50];
int flag;
printf("Write tecplot file (1) or plain text file (0): ");
scanf("%d",&flag);
if ((flag != 1) && (flag != 0)) {
printf("Error: Invalid input. Should be 1 or 0.\n");
return(0);
}
printf("Reading solver.inp.\n");
inputs = fopen("solver.inp","r");
if (!inputs) {
fprintf(stderr,"Error: File \"solver.inp\" not found.\n");
return(1);
} else {
char word[100];
fscanf(inputs,"%s",word);
if (!strcmp(word, "begin")){
while (strcmp(word, "end")){
fscanf(inputs,"%s",word);
if (!strcmp(word, "op_file_format" )) fscanf(inputs,"%s" ,op_file_format);
else if (!strcmp(word, "op_overwrite" )) fscanf(inputs,"%s" ,overwrite );
}
}
fclose(inputs);
}
if (strcmp(op_file_format,"binary") && strcmp(op_file_format,"bin")) {
printf("Error: solution output needs to be in binary files.\n");
return(0);
}
Parameters params;
params.grav_x = 0.0;
params.grav_y = 9.8;
params.R = 287.058;
params.gamma = 1.4;
params.P_ref = 100000.0;
params.rho_ref = 100000.0 / (params.R * 300.0);
params.HB = 0;
printf("Reading physics.inp.\n");
inputs = fopen("physics.inp","r");
if (!inputs) {
fprintf(stderr,"Error: File \"physics.inp\" not found.\n");
return(1);
} else {
char word[100];
fscanf(inputs,"%s",word);
if (!strcmp(word, "begin")){
while (strcmp(word, "end")){
fscanf(inputs,"%s",word);
if (!strcmp(word, "gamma")) fscanf(inputs,"%lf",¶ms.gamma);
else if (!strcmp(word, "gravity")) {
fscanf(inputs,"%lf",¶ms.grav_x);
fscanf(inputs,"%lf",¶ms.grav_y);
} else if (!strcmp(word,"p_ref")) fscanf(inputs,"%lf",¶ms.P_ref);
else if (!strcmp(word,"rho_ref")) fscanf(inputs,"%lf",¶ms.rho_ref);
else if (!strcmp(word,"HB")) fscanf(inputs,"%d",¶ms.HB);
}
}
fclose(inputs);
}
if (params.HB != 2) {
printf("Error: \"HB\" must be specified as 2 in physics.inp.\n");
return(0);
}
if (!strcmp(overwrite,"no")) {
strcpy(filename,"op_fg_00000.bin");
while(1) {
strcpy(tecfile,filename);
tecfile[12] = 'd';
tecfile[13] = 'a';
tecfile[14] = 't';
int err = PostProcess(filename, tecfile, ¶ms, flag);
if (err == -1) {
printf("No more files found. Exiting.\n");
break;
}
IncrementFilename(filename);
}
} else if (!strcmp(overwrite,"yes")) {
strcpy(filename,"op_fg.bin");
strcpy(tecfile,filename);
tecfile[6] = 'd';
tecfile[7] = 'a';
tecfile[8] = 't';
int err = PostProcess(filename, tecfile, ¶ms, flag);
if (err == -1) {
printf("Error: op.bin not found.\n");
return(0);
}
}
return(0);
}
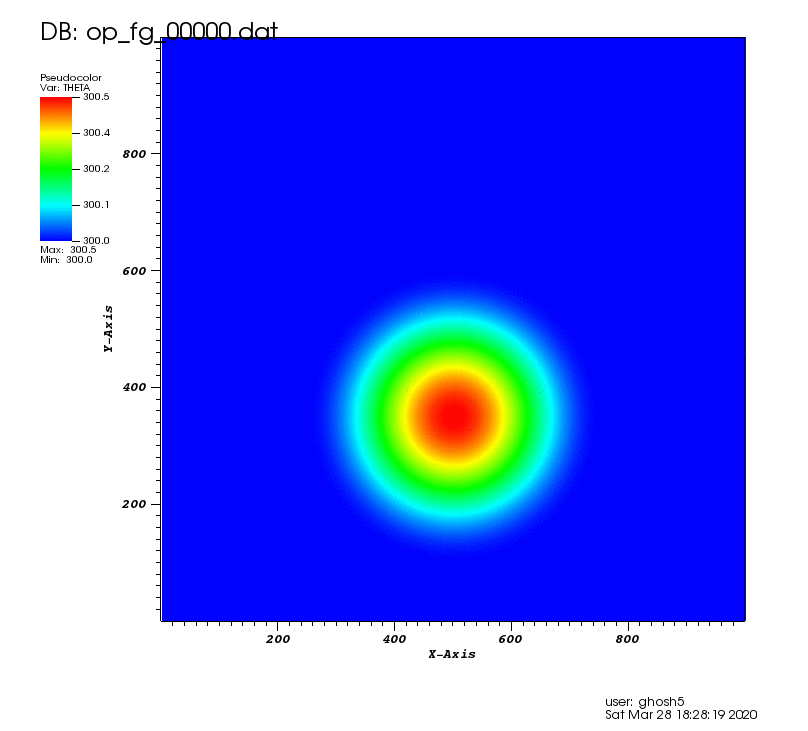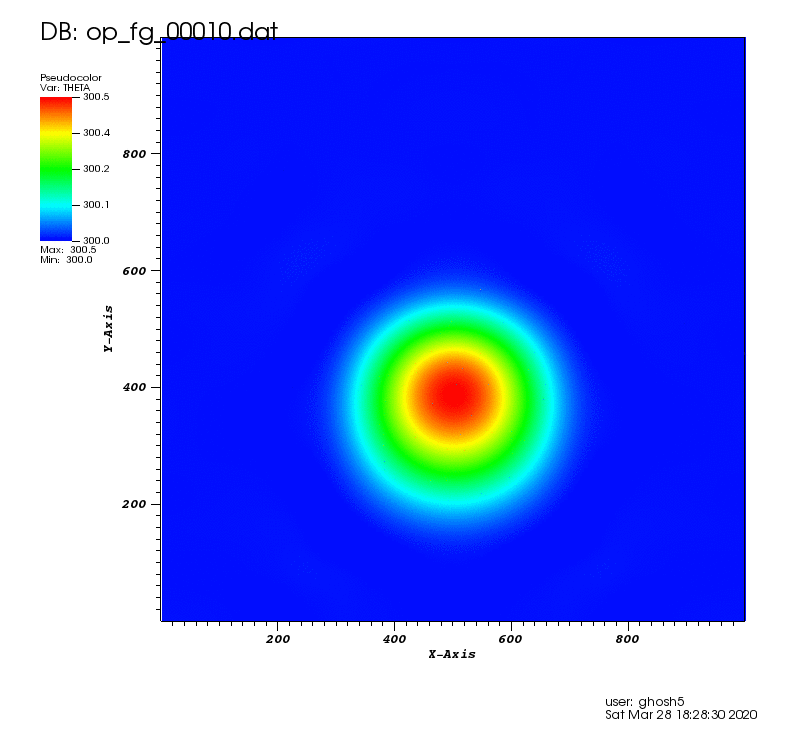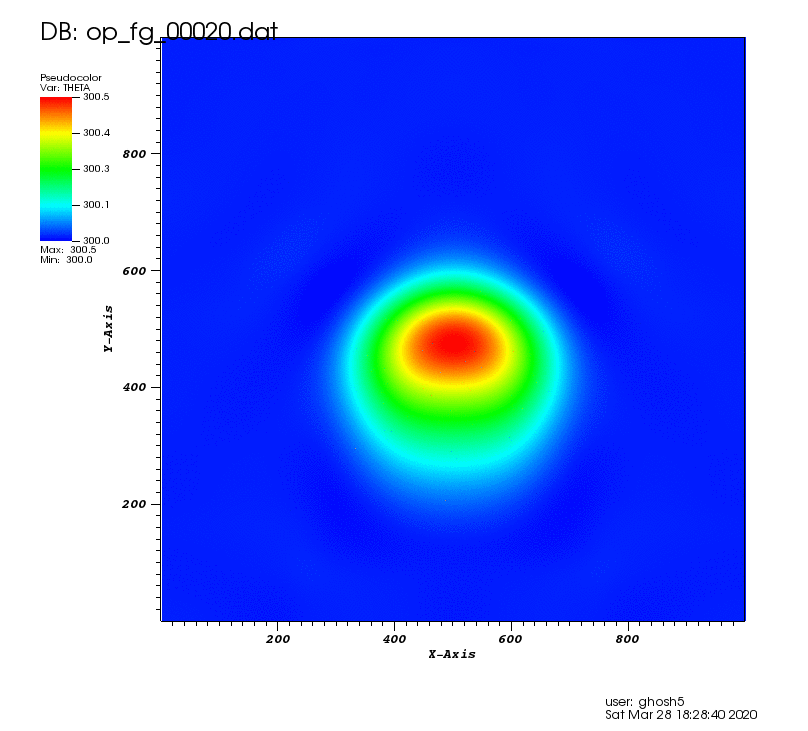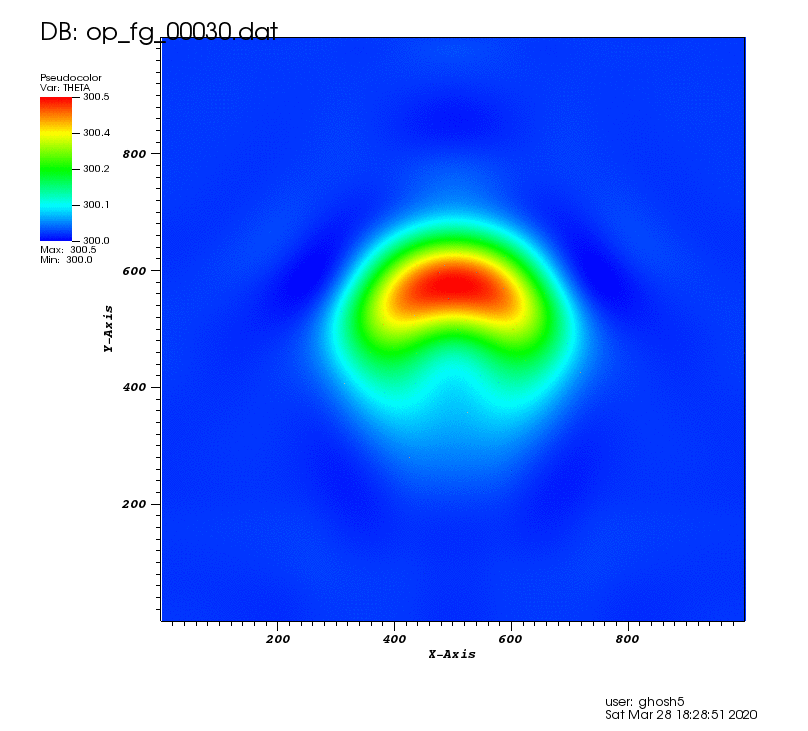
The following animation shows the potential temperature perturbation. It was plotted using VisIt (https://wci.llnl.gov/simulation/computer-codes/visit/) with tecplot2d format chosen in the above postprocessing code.
HyPar - Parallel (MPI) version with 32 processes
Compiled with PETSc time integration.
-- Sparse Grids Simulation --
Sparse grids inputs:
log2 of minimum grid size: 3
interpolation order: 6
write sparse grids solutions? no
Allocated full grid simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 2
No. of variables : 4
Domain size : 256 256
Processes along each dimension : 8 4
No. of ghosts pts : 3
No. of iter. : 30000
Restart iteration : 0
Time integration scheme : rk (ssprk3)
Spatial discretization scheme (hyperbolic) : upw5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : components
Spatial discretization type (parabolic ) : nonconservative-2stage
Spatial discretization scheme (parabolic ) : 4
Time Step : 1.000000E-02
Check for conservation : no
Screen output iterations : 500
File output iterations : 1000
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : no
Physical model : navierstokes2d
Processor distribution for full grid object: 8 4
Initializing sparse grids...
Number of spatial dimensions: 2
Computing sparse grids dimensions...
Number of sparse grid domains in combination technique: 11
Computing processor decompositions...
Sparse Grids: Combination technique grids sizes and coefficients are:-
0: dim = ( 256 8 ), coeff = +1.00e+00, iproc = ( 32 1 )
1: dim = ( 128 16 ), coeff = +1.00e+00, iproc = ( 16 2 )
2: dim = ( 64 32 ), coeff = +1.00e+00, iproc = ( 8 4 )
3: dim = ( 32 64 ), coeff = +1.00e+00, iproc = ( 4 8 )
4: dim = ( 16 128 ), coeff = +1.00e+00, iproc = ( 2 16 )
5: dim = ( 8 256 ), coeff = +1.00e+00, iproc = ( 1 32 )
6: dim = ( 128 8 ), coeff = -1.00e+00, iproc = ( 32 1 )
7: dim = ( 64 16 ), coeff = -1.00e+00, iproc = ( 16 2 )
8: dim = ( 32 32 ), coeff = -1.00e+00, iproc = ( 8 4 )
9: dim = ( 16 64 ), coeff = -1.00e+00, iproc = ( 2 16 )
10: dim = ( 8 128 ), coeff = -1.00e+00, iproc = ( 1 32 )
Allocating data arrays for full grid.
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 1.1234279568688401E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801254424407306E+11
Interpolating grid coordinates to sparse grids domain 0.
Interpolating grid coordinates to sparse grids domain 1.
Interpolating grid coordinates to sparse grids domain 2.
Interpolating grid coordinates to sparse grids domain 3.
Interpolating grid coordinates to sparse grids domain 4.
Interpolating grid coordinates to sparse grids domain 5.
Interpolating grid coordinates to sparse grids domain 6.
Interpolating grid coordinates to sparse grids domain 7.
Interpolating grid coordinates to sparse grids domain 8.
Interpolating grid coordinates to sparse grids domain 9.
Interpolating grid coordinates to sparse grids domain 10.
Domain 0: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 1: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 2: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 3: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 4: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 5: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 6: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 7: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 8: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 9: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Domain 10: Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Initializing solvers.
Initializing physics. Model = "navierstokes2d"
Reading physical model inputs from file "physics.inp".
Interpolating initial solution to sparse grids domain 0.
Volume integral of the initial solution on sparse grids domain 0:
0: 1.1234251586382261E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801105349946042E+11
Interpolating initial solution to sparse grids domain 1.
Volume integral of the initial solution on sparse grids domain 1:
0: 1.1234271613915111E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801217265073633E+11
Interpolating initial solution to sparse grids domain 2.
Volume integral of the initial solution on sparse grids domain 2:
0: 1.1234277731541058E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801245243865408E+11
Interpolating initial solution to sparse grids domain 3.
Volume integral of the initial solution on sparse grids domain 3:
0: 1.1234449109078781E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801756102382306E+11
Interpolating initial solution to sparse grids domain 4.
Volume integral of the initial solution on sparse grids domain 4:
0: 1.1233656900511470E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3799406404289981E+11
Interpolating initial solution to sparse grids domain 5.
Volume integral of the initial solution on sparse grids domain 5:
0: 1.1234285119873849E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801254424407306E+11
Interpolating initial solution to sparse grids domain 6.
Volume integral of the initial solution on sparse grids domain 6:
0: 1.1234251586382259E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801105349946048E+11
Interpolating initial solution to sparse grids domain 7.
Volume integral of the initial solution on sparse grids domain 7:
0: 1.1234271612869459E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801217265073633E+11
Interpolating initial solution to sparse grids domain 8.
Volume integral of the initial solution on sparse grids domain 8:
0: 1.1234277774468665E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801245243865405E+11
Interpolating initial solution to sparse grids domain 9.
Volume integral of the initial solution on sparse grids domain 9:
0: 1.1234449553994713E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3801756102382306E+11
Interpolating initial solution to sparse grids domain 10.
Volume integral of the initial solution on sparse grids domain 10:
0: 1.1233661938574852E+06
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 2.3799406404289972E+11
Setting up time integration.
Solving in time (from 0 to 30000 iterations)
Writing solution file op_fg_00000.bin.
--
Iteration: 500, Time: 5.000e+00
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 6.5813E+00
--
--
Iteration: 1000, Time: 1.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 5.5077E+00
--
Writing solution file op_fg_00001.bin.
--
Iteration: 1500, Time: 1.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 4.9858E+00
--
--
Iteration: 2000, Time: 2.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 4.5788E+00
--
Writing solution file op_fg_00002.bin.
--
Iteration: 2500, Time: 2.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 4.2167E+00
--
--
Iteration: 3000, Time: 3.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.9285E+00
--
Writing solution file op_fg_00003.bin.
--
Iteration: 3500, Time: 3.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 9.9974E-01
--
--
Iteration: 4000, Time: 4.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.6060E+00
--
Writing solution file op_fg_00004.bin.
--
Iteration: 4500, Time: 4.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.5391E+00
--
--
Iteration: 5000, Time: 5.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.4880E+00
--
Writing solution file op_fg_00005.bin.
--
Iteration: 5500, Time: 5.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.3799E+00
--
--
Iteration: 6000, Time: 6.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.2350E+00
--
Writing solution file op_fg_00006.bin.
--
Iteration: 6500, Time: 6.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 3.1020E+00
--
--
Iteration: 7000, Time: 7.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 7.8745E-01
--
Writing solution file op_fg_00007.bin.
--
Iteration: 7500, Time: 7.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.9683E+00
--
--
Iteration: 8000, Time: 8.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.9582E+00
--
Writing solution file op_fg_00008.bin.
--
Iteration: 8500, Time: 8.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.9630E+00
--
--
Iteration: 9000, Time: 9.000e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.8798E+00
--
Writing solution file op_fg_00009.bin.
--
Iteration: 9500, Time: 9.500e+01
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.7983E+00
--
--
Iteration: 10000, Time: 1.000e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.7053E+00
--
Writing solution file op_fg_00010.bin.
--
Iteration: 10500, Time: 1.050e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 7.4930E-01
--
--
Iteration: 11000, Time: 1.100e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.6256E+00
--
Writing solution file op_fg_00011.bin.
--
Iteration: 11500, Time: 1.150e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.6302E+00
--
--
Iteration: 12000, Time: 1.200e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.6453E+00
--
Writing solution file op_fg_00012.bin.
--
Iteration: 12500, Time: 1.250e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.5970E+00
--
--
Iteration: 13000, Time: 1.300e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.5272E+00
--
Writing solution file op_fg_00013.bin.
--
Iteration: 13500, Time: 1.350e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.4529E+00
--
--
Iteration: 14000, Time: 1.400e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 7.8316E-01
--
Writing solution file op_fg_00014.bin.
--
Iteration: 14500, Time: 1.450e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.3973E+00
--
--
Iteration: 15000, Time: 1.500e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.4074E+00
--
Writing solution file op_fg_00015.bin.
--
Iteration: 15500, Time: 1.550e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.4236E+00
--
--
Iteration: 16000, Time: 1.600e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.4106E+00
--
Writing solution file op_fg_00016.bin.
--
Iteration: 16500, Time: 1.650e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.3345E+00
--
--
Iteration: 17000, Time: 1.700e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.2712E+00
--
Writing solution file op_fg_00017.bin.
--
Iteration: 17500, Time: 1.750e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 8.4438E-01
--
--
Iteration: 18000, Time: 1.800e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.2288E+00
--
Writing solution file op_fg_00018.bin.
--
Iteration: 18500, Time: 1.850e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.2418E+00
--
--
Iteration: 19000, Time: 1.900e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.2576E+00
--
Writing solution file op_fg_00019.bin.
--
Iteration: 19500, Time: 1.950e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.2643E+00
--
--
Iteration: 20000, Time: 2.000e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.1870E+00
--
Writing solution file op_fg_00020.bin.
--
Iteration: 20500, Time: 2.050e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.1311E+00
--
--
Iteration: 21000, Time: 2.100e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 9.1303E-01
--
Writing solution file op_fg_00021.bin.
--
Iteration: 21500, Time: 2.150e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0969E+00
--
--
Iteration: 22000, Time: 2.200e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.1118E+00
--
Writing solution file op_fg_00022.bin.
--
Iteration: 22500, Time: 2.250e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.1269E+00
--
--
Iteration: 23000, Time: 2.300e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.1371E+00
--
Writing solution file op_fg_00023.bin.
--
Iteration: 23500, Time: 2.350e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0685E+00
--
--
Iteration: 24000, Time: 2.400e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0180E+00
--
Writing solution file op_fg_00024.bin.
--
Iteration: 24500, Time: 2.450e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 9.8161E-01
--
--
Iteration: 25000, Time: 2.500e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9894E+00
--
Writing solution file op_fg_00025.bin.
--
Iteration: 25500, Time: 2.550e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0059E+00
--
--
Iteration: 26000, Time: 2.600e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0204E+00
--
Writing solution file op_fg_00026.bin.
--
Iteration: 26500, Time: 2.650e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 2.0221E+00
--
--
Iteration: 27000, Time: 2.700e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9700E+00
--
Writing solution file op_fg_00027.bin.
--
Iteration: 27500, Time: 2.750e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9239E+00
--
--
Iteration: 28000, Time: 2.800e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.0484E+00
--
Writing solution file op_fg_00028.bin.
--
Iteration: 28500, Time: 2.850e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.8991E+00
--
--
Iteration: 29000, Time: 2.900e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9173E+00
--
Writing solution file op_fg_00029.bin.
--
Iteration: 29500, Time: 2.950e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9311E+00
--
--
Iteration: 30000, Time: 3.000e+02
Max CFL: 4.427E-01, Max Diff. No.: -1.000E+00, Norm: 1.9177E+00
--
Writing solution file op_fg_00030.bin.
Completed time integration (Final time: 300.000000).
Solver runtime (in seconds): 6.0051986999999997E+01
Total runtime (in seconds): 6.0287636999999997E+01
Deallocating arrays.
Finished.