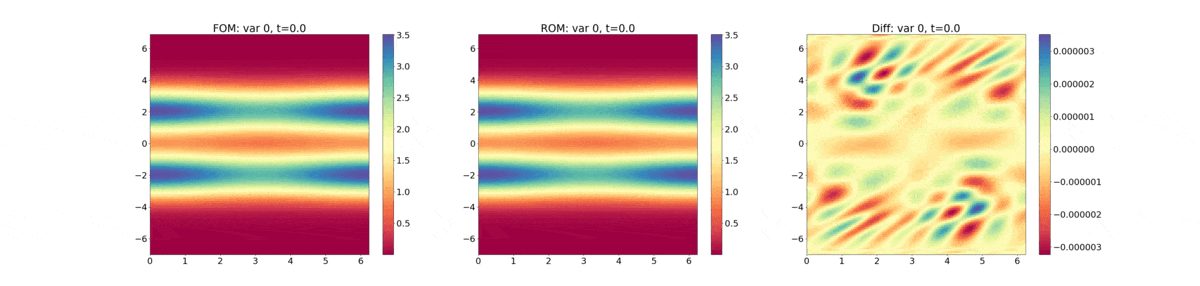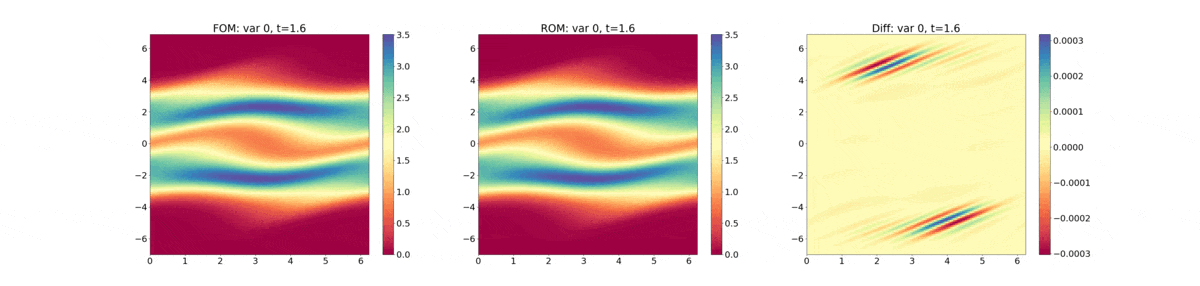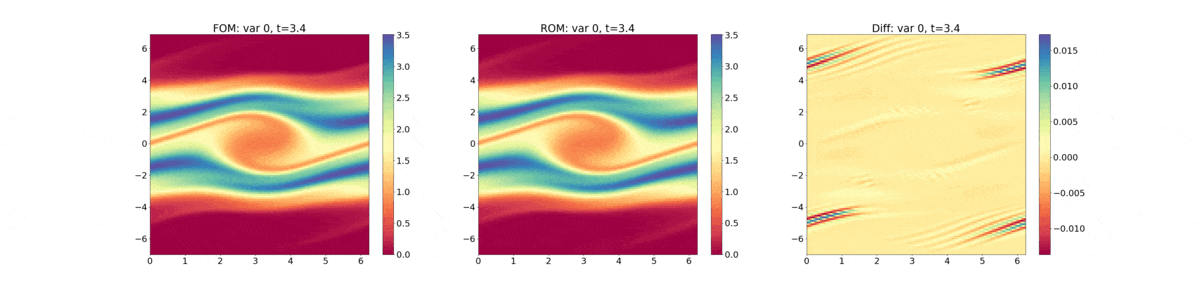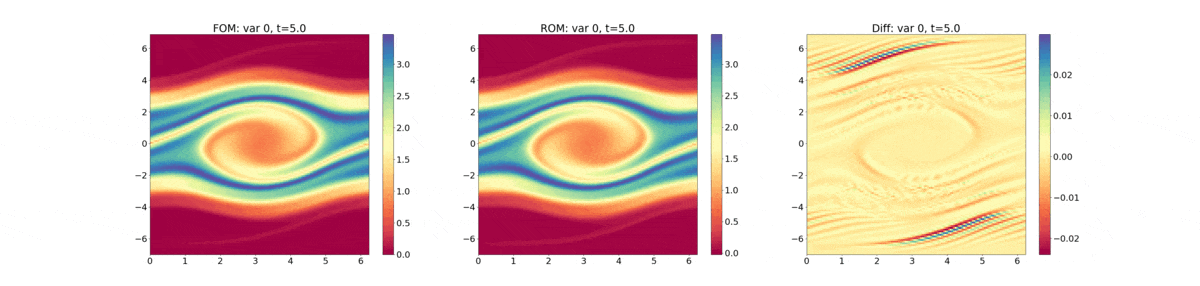
Reduced Order Modeling: This example trains a DMD object and then predicts the solution using the DMD at the same times that the actual HyPar solution is written at.
Initial solution: \(f\left(x,v\right) = \frac{4}{\pi T}\left(1+\frac{1}{10}\cos\left(2k\pi\frac{x}{L}\right)\right)\left[\exp\left(-\frac{\left(v-2\right)^2}{2T}\right) + \exp\left(-\frac{\left(v+2\right)^2}{2T}\right)\right]\), \(k=1,T=1,L=2\pi\).
Self-consistent electric field is computed by solving the Poisson equation in a periodic domain using Fourier transforms. This examples requires HyPar to be compiled with FFTW (http://www.fftw.org/).
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
{
double pi = 4.0 * atan(1.0);
double two_pi = 8.0 * atan(1.0);
double L = two_pi;
double T = 1.0;
double k = 1.0;
int NI,NJ,ndims,n_iter;
FILE *in;
char ip_file_type[50];
strcpy(ip_file_type,"ascii");
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) {
fprintf(stderr,"Error: Input file \"solver.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")){
while (strcmp(word, "end")){
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) fscanf(in,"%d",&ndims);
else if (!strcmp(word, "size")) {
fscanf(in,"%d",&NI);
fscanf(in,"%d",&NJ);
} else if (!strcmp(word, "ip_file_type")) {
fscanf(in,"%s",ip_file_type);
}
}
} else {
fprintf(stderr,"Error: Illegal format in solver.inp. Crash and burn!\n");
return(0);
}
}
fclose(in);
if (ndims != 2) {
fprintf(stderr,"ndims is not 2 in solver.inp. this code is to generate 2D initial conditions\n");
return(0);
}
printf("Grid: %d, %d\n",NI,NJ);
int i,j;
double dx = two_pi / ((double)NI);
double dv = (14.0 * T) / ((double)NJ);
double start_x = 0.0;
double start_v = -7.0 * T;
double *x, *v, *f;
x = (double*) calloc (NI , sizeof(double));
v = (double*) calloc (NJ , sizeof(double));
f = (double*) calloc (NI*NJ, sizeof(double));
for (i = 0; i < NI; i++){
for (j = 0; j < NJ; j++){
x[i] = start_x + i*dx;
v[j] = start_v + j*dv;
int p = NJ*i + j;
double temp1 = cos(2.0 * k * pi * x[i] / L);
double temp2 = exp(- (v[j] -2.0) * (v[j] - 2.0) / (2.0 * T));
double temp3 = exp(- (v[j] + 2.0) * (v[j] + 2.0) / (2.0 * T));
double temp4 = sqrt(2.0 * pi * T);
f[p] = 16.0 * (1.0 + 0.1 * temp1) * 0.5 * (temp2 / temp4 + temp3 / temp4);
}
}
FILE *out;
if (!strcmp(ip_file_type,"ascii")) {
out = fopen("initial.inp","w");
for (i = 0; i < NI; i++) fprintf(out,"%1.16e ",x[i]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) fprintf(out,"%1.16e ",v[j]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%1.16e ", f[p]);
}
}
fprintf(out,"\n");
fclose(out);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary exact solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x, sizeof(double),NI,out);
fwrite(v, sizeof(double),NJ,out);
double *F = (double*) calloc (NI*NJ,sizeof(double));
for (i=0; i < NI; i++) {
for (j = 0; j < NJ; j++) {
int p = NJ*i + j;
int q = NI*j + i;
F[q+0] = f[p];
}
}
fwrite(F, sizeof(double),NI*NJ,out);
free(F);
fclose(out);
}
free(x);
free(v);
free(f);
return(0);
}
The first of each of these file sets is the solution at \(t=0\) and the final one is the solution at \(t=5\). Since HyPar::op_overwrite is set to no in solver.inp, a separate file is written for solutions at each output time. All the files are binary (HyPar::op_file_format is set to binary in solver.inp).
The following plot shows the evolution of the distribution function - FOM (full-order model) refers to the HyPar solution, ROM (reduced-order model) refers to the DMD solution, and Diff is the difference between the two.
By default, the code will write the trained DMD object(s) to files in a subdirectory (DMDROMObject::m_dirname - default value is "DMD"). If the subdirectory does not exist, the code may not report an error (or give some HDF5 file-writing error); the DMD objects will not be written! If the subdirectory exists, several files will exist after the simulation is complete - they are in a format that is readable by libROM.
HyPar - Parallel (MPI) version with 16 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 2
No. of variables : 1
Domain size : 128 128
Processes along each dimension : 4 4
Exact solution domain size : 128 128
No. of ghosts pts : 3
No. of iter. : 1000
Restart iteration : 0
Time integration scheme : rk (44)
Spatial discretization scheme (hyperbolic) : weno5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : characteristic
Spatial discretization type (parabolic ) : nonconservative-1stage
Spatial discretization scheme (parabolic ) : 2
Time Step : 5.000000E-03
Check for conservation : yes
Screen output iterations : 20
File output iterations : 40
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : no
Physical model : vlasov
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 1.0053093535576996E+02
Reading boundary conditions from boundary.inp.
Boundary periodic: Along dimension 0 and face +1
Boundary periodic: Along dimension 0 and face -1
Boundary dirichlet: Along dimension 1 and face +1
Boundary dirichlet: Along dimension 1 and face -1
4 boundary condition(s) read.
Initializing solvers.
Reading WENO parameters from weno.inp.
Initializing physics. Model = "vlasov"
Reading physical model inputs from file "physics.inp".
Setting up time integration.
Setting up libROM interface.
libROM inputs and parameters:
reduced model dimensionality: 16
sampling frequency: 1
mode: train
type: DMD
save to file: true
local vector size: 1024
libROM DMD inputs:
number of samples per window: 200
directory name for DMD onjects: DMD
Solving in time (from 0 to 1000 iterations)
Writing solution file op_00000.bin.
DMDROMObject::takeSample() - creating new DMD object, t=0.000000 (total: 1).
iter= 20 t=1.000E-01 CFL=7.019E-01 norm=5.9995E-03 wctime: 1.1E-02 (s) cons_err=9.8951E-16
iter= 40 t=2.000E-01 CFL=7.019E-01 norm=4.5861E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00001.bin.
iter= 60 t=3.000E-01 CFL=7.019E-01 norm=3.5020E-03 wctime: 1.1E-02 (s) cons_err=5.6543E-16
iter= 80 t=4.000E-01 CFL=7.019E-01 norm=4.1353E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00002.bin.
iter= 100 t=5.000E-01 CFL=7.019E-01 norm=5.3650E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
iter= 120 t=6.000E-01 CFL=7.019E-01 norm=5.8305E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00003.bin.
iter= 140 t=7.000E-01 CFL=7.019E-01 norm=5.2115E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 160 t=8.000E-01 CFL=7.019E-01 norm=3.9852E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
Writing solution file op_00004.bin.
iter= 180 t=9.000E-01 CFL=7.019E-01 norm=3.4521E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 200 t=1.000E+00 CFL=7.019E-01 norm=4.2749E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00005.bin.
DMDROMObject::takeSample() - creating new DMD object, t=1.000000 (total: 2).
iter= 220 t=1.100E+00 CFL=7.019E-01 norm=5.1552E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 240 t=1.200E+00 CFL=7.019E-01 norm=5.1908E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
Writing solution file op_00006.bin.
iter= 260 t=1.300E+00 CFL=7.019E-01 norm=4.3530E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
iter= 280 t=1.400E+00 CFL=7.019E-01 norm=3.3849E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00007.bin.
iter= 300 t=1.500E+00 CFL=7.019E-01 norm=3.4785E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 320 t=1.600E+00 CFL=7.019E-01 norm=4.3003E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00008.bin.
iter= 340 t=1.700E+00 CFL=7.019E-01 norm=4.6344E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 360 t=1.800E+00 CFL=7.019E-01 norm=4.0634E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00009.bin.
iter= 380 t=1.900E+00 CFL=7.019E-01 norm=3.0361E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 400 t=2.000E+00 CFL=7.019E-01 norm=2.9697E-03 wctime: 1.1E-02 (s) cons_err=5.6543E-16
Writing solution file op_00010.bin.
DMDROMObject::takeSample() - creating new DMD object, t=2.000000 (total: 3).
iter= 420 t=2.100E+00 CFL=7.019E-01 norm=4.1787E-03 wctime: 1.1E-02 (s) cons_err=7.0679E-16
iter= 440 t=2.200E+00 CFL=7.019E-01 norm=5.2329E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00011.bin.
iter= 460 t=2.300E+00 CFL=7.019E-01 norm=5.4099E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
iter= 480 t=2.400E+00 CFL=7.019E-01 norm=4.7395E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00012.bin.
iter= 500 t=2.500E+00 CFL=7.019E-01 norm=3.9095E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 520 t=2.600E+00 CFL=7.019E-01 norm=3.9147E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00013.bin.
iter= 540 t=2.700E+00 CFL=7.019E-01 norm=4.5873E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
iter= 560 t=2.800E+00 CFL=7.019E-01 norm=4.9026E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
Writing solution file op_00014.bin.
iter= 580 t=2.900E+00 CFL=7.019E-01 norm=4.3939E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 600 t=3.000E+00 CFL=7.019E-01 norm=3.3860E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00015.bin.
DMDROMObject::takeSample() - creating new DMD object, t=3.000000 (total: 4).
iter= 620 t=3.100E+00 CFL=7.019E-01 norm=3.1040E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 640 t=3.200E+00 CFL=7.019E-01 norm=4.1295E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
Writing solution file op_00016.bin.
iter= 660 t=3.300E+00 CFL=7.019E-01 norm=5.2050E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 680 t=3.400E+00 CFL=7.019E-01 norm=5.5118E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00017.bin.
iter= 700 t=3.500E+00 CFL=7.019E-01 norm=5.0084E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 720 t=3.600E+00 CFL=7.019E-01 norm=4.3083E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
Writing solution file op_00018.bin.
iter= 740 t=3.700E+00 CFL=7.019E-01 norm=4.3217E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
iter= 760 t=3.800E+00 CFL=7.019E-01 norm=4.9976E-03 wctime: 1.3E-02 (s) cons_err=0.0000E+00
Writing solution file op_00019.bin.
iter= 780 t=3.900E+00 CFL=7.019E-01 norm=5.4117E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
iter= 800 t=4.000E+00 CFL=7.019E-01 norm=5.0478E-03 wctime: 1.1E-02 (s) cons_err=5.6543E-16
Writing solution file op_00020.bin.
DMDROMObject::takeSample() - creating new DMD object, t=4.000000 (total: 5).
iter= 820 t=4.100E+00 CFL=7.019E-01 norm=4.1065E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
iter= 840 t=4.200E+00 CFL=7.019E-01 norm=3.5134E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00021.bin.
iter= 860 t=4.300E+00 CFL=7.019E-01 norm=4.0055E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
iter= 880 t=4.400E+00 CFL=7.019E-01 norm=4.8258E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
Writing solution file op_00022.bin.
iter= 900 t=4.500E+00 CFL=7.019E-01 norm=5.1295E-03 wctime: 1.1E-02 (s) cons_err=5.6543E-16
iter= 920 t=4.600E+00 CFL=7.019E-01 norm=4.6994E-03 wctime: 1.1E-02 (s) cons_err=1.4136E-16
Writing solution file op_00023.bin.
iter= 940 t=4.700E+00 CFL=7.019E-01 norm=3.9034E-03 wctime: 1.1E-02 (s) cons_err=4.2407E-16
iter= 960 t=4.800E+00 CFL=7.019E-01 norm=3.6356E-03 wctime: 1.1E-02 (s) cons_err=2.8272E-16
Writing solution file op_00024.bin.
iter= 980 t=4.900E+00 CFL=7.019E-01 norm=4.2720E-03 wctime: 1.1E-02 (s) cons_err=5.6543E-16
iter= 1000 t=5.000E+00 CFL=7.019E-01 norm=4.9911E-03 wctime: 1.1E-02 (s) cons_err=0.0000E+00
Completed time integration (Final time: 5.000000), total wctime: 11.340854 (seconds).
Writing solution file op_00025.bin.
libROM: Training ROM.
DMDROMObject::train() - training DMD object 0 with 201 samples.
Using 16 basis vectors out of 200.
DMDROMObject::train() - training DMD object 1 with 201 samples.
Using 16 basis vectors out of 200.
DMDROMObject::train() - training DMD object 2 with 201 samples.
Using 16 basis vectors out of 200.
DMDROMObject::train() - training DMD object 3 with 201 samples.
Using 16 basis vectors out of 200.
DMDROMObject::train() - training DMD object 4 with 200 samples.
Using 16 basis vectors out of 199.
libROM: total training wallclock time: 2.298255 (seconds).
libROM: Predicting solution at time 0.0000e+00 using ROM.
libROM: wallclock time: 0.013076 (seconds).
Writing solution file op_rom_00000.bin.
libROM: Predicting solution at time 2.0000e-01 using ROM.
libROM: wallclock time: 0.012934 (seconds).
Writing solution file op_rom_00001.bin.
libROM: Predicting solution at time 4.0000e-01 using ROM.
libROM: wallclock time: 0.012966 (seconds).
Writing solution file op_rom_00002.bin.
libROM: Predicting solution at time 6.0000e-01 using ROM.
libROM: wallclock time: 0.012916 (seconds).
Writing solution file op_rom_00003.bin.
libROM: Predicting solution at time 8.0000e-01 using ROM.
libROM: wallclock time: 0.012921 (seconds).
Writing solution file op_rom_00004.bin.
libROM: Predicting solution at time 1.0000e+00 using ROM.
libROM: wallclock time: 0.012934 (seconds).
Writing solution file op_rom_00005.bin.
libROM: Predicting solution at time 1.2000e+00 using ROM.
libROM: wallclock time: 0.012907 (seconds).
Writing solution file op_rom_00006.bin.
libROM: Predicting solution at time 1.4000e+00 using ROM.
libROM: wallclock time: 0.012912 (seconds).
Writing solution file op_rom_00007.bin.
libROM: Predicting solution at time 1.6000e+00 using ROM.
libROM: wallclock time: 0.012952 (seconds).
Writing solution file op_rom_00008.bin.
libROM: Predicting solution at time 1.8000e+00 using ROM.
libROM: wallclock time: 0.012914 (seconds).
Writing solution file op_rom_00009.bin.
libROM: Predicting solution at time 2.0000e+00 using ROM.
libROM: wallclock time: 0.013504 (seconds).
Writing solution file op_rom_00010.bin.
libROM: Predicting solution at time 2.2000e+00 using ROM.
libROM: wallclock time: 0.013461 (seconds).
Writing solution file op_rom_00011.bin.
libROM: Predicting solution at time 2.4000e+00 using ROM.
libROM: wallclock time: 0.013477 (seconds).
Writing solution file op_rom_00012.bin.
libROM: Predicting solution at time 2.6000e+00 using ROM.
libROM: wallclock time: 0.013462 (seconds).
Writing solution file op_rom_00013.bin.
libROM: Predicting solution at time 2.8000e+00 using ROM.
libROM: wallclock time: 0.013809 (seconds).
Writing solution file op_rom_00014.bin.
libROM: Predicting solution at time 3.0000e+00 using ROM.
libROM: wallclock time: 0.014043 (seconds).
Writing solution file op_rom_00015.bin.
libROM: Predicting solution at time 3.2000e+00 using ROM.
libROM: wallclock time: 0.013693 (seconds).
Writing solution file op_rom_00016.bin.
libROM: Predicting solution at time 3.4000e+00 using ROM.
libROM: wallclock time: 0.013528 (seconds).
Writing solution file op_rom_00017.bin.
libROM: Predicting solution at time 3.6000e+00 using ROM.
libROM: wallclock time: 0.013531 (seconds).
Writing solution file op_rom_00018.bin.
libROM: Predicting solution at time 3.8000e+00 using ROM.
libROM: wallclock time: 0.012887 (seconds).
Writing solution file op_rom_00019.bin.
libROM: Predicting solution at time 4.0000e+00 using ROM.
libROM: wallclock time: 0.012988 (seconds).
Writing solution file op_rom_00020.bin.
libROM: Predicting solution at time 4.2000e+00 using ROM.
libROM: wallclock time: 0.012888 (seconds).
Writing solution file op_rom_00021.bin.
libROM: Predicting solution at time 4.4000e+00 using ROM.
libROM: wallclock time: 0.012920 (seconds).
Writing solution file op_rom_00022.bin.
libROM: Predicting solution at time 4.6000e+00 using ROM.
libROM: wallclock time: 0.013062 (seconds).
Writing solution file op_rom_00023.bin.
libROM: Predicting solution at time 4.8000e+00 using ROM.
libROM: wallclock time: 0.013269 (seconds).
Writing solution file op_rom_00024.bin.
libROM: Predicting solution at time 5.0000e+00 using ROM.
libROM: wallclock time: 0.013103 (seconds).
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom_00025.bin.
libROM: total prediction/query wallclock time: 0.343057 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object with filename root DMD/dmdobj_0000.
Saving DMD object with filename root DMD/dmdobj_0001.
Saving DMD object with filename root DMD/dmdobj_0002.
Saving DMD object with filename root DMD/dmdobj_0003.
Saving DMD object with filename root DMD/dmdobj_0004.
Conservation Errors:
0.0000000000000000E+00
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 1.9745400769612493E-03
L2 Norm : 2.4778613956876021E-03
Linfinity Norm : 8.6020754178481092E-03
Solver runtime (in seconds): 2.2483938999999999E+01
Total runtime (in seconds): 2.3051815999999999E+01
Deallocating arrays.
Finished.