See 2D Navier-Stokes Equations - Lid-Driven Square Cavity to familiarize yourself with this case.
Location: hypar/Examples/2D/NavierStokes2D/LidDrivenCavity_libROM_DMD_Train (This directory contains all the input files needed to run this case.)
Governing equations: 2D Navier-Stokes Equations (navierstokes2d.h)
Reduced Order Modeling: This example trains a DMD object and then predicts the solution using the DMD at the same times that the actual HyPar solution is written at.
Reference:
- Erturk, E., Corke, T.C., and Gokcol, C., "Numerical Solutions of
2-D Steady Incompressible Driven Cavity Flow at High Reynolds Numbers", International Journal for Numerical Methods in Fluids, 48, 2005, http://dx.doi.org/10.1002/fld.953.
- Ghia, U., Ghia, K.N., Shin, C.T., "High-Re Solutions for Incompressible
Flow using the Navier-Stokes Equations and a Multigrid Method", Journal of Computational Physics, 48, 1982, http://dx.doi.org/10.1016/0021-9991(82)90058-4.
Note that this is an incompressible problem being solved here using the compressible Navier-Stokes equations in terms of non-dimensional flow variables. The density and pressure are taken such that the speed of sound is 1.0, and the flow velocities specified in the initial and boundary conditions correspond to a characteristic Mach number of 0.1 (thus, they are 0.1 times the values in the above reference).
Domain: \(0 \le x, y \le 1\)
Boundary conditions:
- No-slip wall BC on \(x=0,1, 0 \le y \le 1\) (_NOSLIP_WALL_ with 0 wall velocity).
- No-slip wall BC on \(y=0, 0 \le x \le 1\) (_NOSLIP_WALL_ with 0 wall velocity).
- Moving no-slip wall BC on \(y=1, 0 \le x \le 1\) (_NOSLIP_WALL_ with specified wall velocity of 0.1 in the x-direction).
Initial solution: \(\rho=1, p=1/\gamma\). The velocities are specified according to the references above, but scaled by a factor of 0.1 to ensure that the characteristic Mach number is 0.1.
Other parameters:
Note: Pressure is taken as \(1/\gamma\) in the above so that the freestream speed of sound is 1.
Numerical method:
Reduced Order Modeling:
Input files required:
librom.inp
begin
rdim 16
sampling_frequency 50
mode train
dmd_num_win_samples 200
end
solver.inp
begin
ndims 2
nvars 4
size 128 128
iproc 4 4
ghost 3
n_iter 50000
time_scheme rk
time_scheme_type 44
hyp_space_scheme upw5
hyp_flux_split no
hyp_interp_type components
par_space_type nonconservative-2stage
par_space_scheme 4
dt 0.005
screen_op_iter 1000
file_op_iter 10000
input_mode serial
ip_file_type binary
output_mode serial
op_file_format binary
op_overwrite yes
model navierstokes2d
end
boundary.inp
4
noslip-wall 0 1 0 0 0 1.0
0.0 0.0
noslip-wall 0 -1 0 0 0 1.0
0.0 0.0
noslip-wall 1 1 0 1.0 0 0
0.0 0.0
noslip-wall 1 -1 0 1.0 0 0
0.1 0.0
physics.inp (Note: this file specifies \(Re = 3200\), change Re here for other Reynolds numbers.)
begin
gamma 1.4
upwinding roe
Pr 0.72
Minf 0.1
Re 3200
end
To generate initial.inp (initial solution), compile and run the following code in the run directory
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
{
double gamma = 1.4;
int NI,NJ,ndims;
char ip_file_type[50]; strcpy(ip_file_type,"ascii");
FILE *in;
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) {
printf("Error: Input file \"solver.inp\" not found. Default values will be used.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")) {
while (strcmp(word, "end")) {
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) fscanf(in,"%d",&ndims);
else if (!strcmp(word, "size")) {
fscanf(in,"%d",&NI);
fscanf(in,"%d",&NJ);
} else if (!strcmp(word, "ip_file_type")) fscanf(in,"%s",ip_file_type);
}
} else printf("Error: Illegal format in solver.inp. Crash and burn!\n");
}
fclose(in);
if (ndims != 2) {
printf("ndims is not 2 in solver.inp. this code is to generate 2D initial conditions\n");
return(0);
}
printf("Grid:\t\t\t%d X %d\n",NI,NJ);
int i,j;
double dx = 1.0 / ((double)NI-1);
double dy = 1.0 / ((double)NJ-1);
double Minf = 0.1;
double *x, *y, *u0, *u1, *u2, *u3;
x = (double*) calloc (NI , sizeof(double));
y = (double*) calloc (NJ , sizeof(double));
u0 = (double*) calloc (NI*NJ, sizeof(double));
u1 = (double*) calloc (NI*NJ, sizeof(double));
u2 = (double*) calloc (NI*NJ, sizeof(double));
u3 = (double*) calloc (NI*NJ, sizeof(double));
for (i = 0; i < NI; i++){
for (j = 0; j < NJ; j++){
x[i] = i*dx;
y[j] = j*dy;
int p = NJ*i + j;
double rho, u, v, P;
rho = 1.0;
P = 1.0/gamma;
u = Minf * (y[j]-0.5);
v = - Minf * (x[i]-0.5);
u0[p] = rho;
u1[p] = rho*u;
u2[p] = rho*v;
u3[p] = P/(gamma-1.0) + 0.5*rho*(u*u+v*v);
}
}
FILE *out;
if (!strcmp(ip_file_type,"ascii")) {
printf("Writing ASCII initial solution file initial.inp\n");
out = fopen("initial.inp","w");
for (i = 0; i < NI; i++) fprintf(out,"%lf ",x[i]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) fprintf(out,"%lf ",y[j]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u0[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u1[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u2[p]);
}
}
fprintf(out,"\n");
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = NJ*i + j;
fprintf(out,"%lf ",u3[p]);
}
}
fprintf(out,"\n");
fclose(out);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary initial solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x,sizeof(double),NI,out);
fwrite(y,sizeof(double),NJ,out);
double *U = (double*) calloc (4*NI*NJ,sizeof(double));
for (i=0; i < NI; i++) {
for (j = 0; j < NJ; j++) {
int p = NJ*i + j;
int q = NI*j + i;
U[4*q+0] = u0[p];
U[4*q+1] = u1[p];
U[4*q+2] = u2[p];
U[4*q+3] = u3[p];
}
}
fwrite(U,sizeof(double),4*NI*NJ,out);
free(U);
fclose(out);
}
free(x);
free(y);
free(u0);
free(u1);
free(u2);
free(u3);
return(0);
}
Output:
Note that iproc is set to
4 4
in solver.inp (i.e., 4 processors along x, and 4 processor along y). Thus, this example should be run with 16 MPI ranks (or change iproc).
After running the code, there should be the following output files:
- 1 output file op.bin; this is the HyPar solutions.
- 1 output file op_rom.bin; this is the predicted solutions from the DMD object(s).
All the files are binary (HyPar::op_file_format is set to binary in solver.inp).
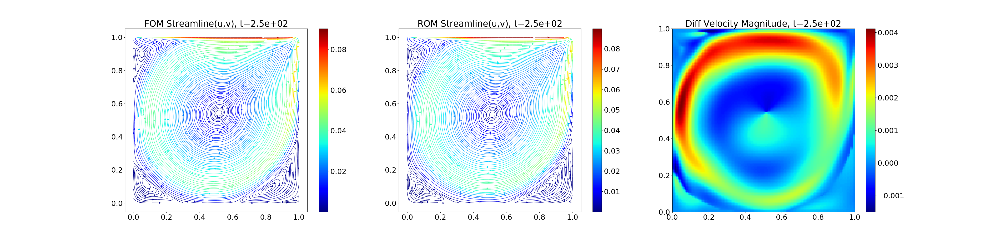
The provided Python script (plotStreamlines.py) can be used to generate plots from the binary files that compare the HyPar and DMD solutions. Alternatively, HyPar::op_file_format can be set to tecplot2d, and Tecplot/VisIt or something similar can be used to plot the resulting text files.
The following plot shows the final solution (velocity streamlines colored by the velocity magnitude) - FOM (full-order model) refers to the HyPar solution, ROM (reduced-order model) refers to the DMD solution, and Diff is the difference in the velocity magnitude between the two.
Wall clock times:
- PDE solution: 554.6 seconds
- DMD training time: 58.6 seconds
- DMD prediction/query time: 0.3 seconds
The L1, L2, and Linf norms of the diff between the HyPar and ROM solution at the final time are calculated and reported on screen (see below) as well as pde_rom_diff.dat:
128 128 4 4 5.0000000000000001E-03 1.7418241509090257E-02 1.7510056779346722E-02 1.8794021053187084E-02 5.7135094100000003E+02 5.7163283200000001E+02
The numbers are: number of grid points in each dimension (HyPar::dim_global), number of processors in each dimension (MPIVariables::iproc), time step size (HyPar::dt), L1, L2, and L-infinity norms of the diff (HyPar::rom_diff_norms), solver wall time (seconds) (i.e., not accounting for initialization, and cleaning up), and total wall time.
By default, the code will write the trained DMD object(s) to files in a subdirectory (DMDROMObject::m_dirname - default value is "DMD"). If the subdirectory does not exist, the code may not report an error (or give some HDF5 file-writing error); the DMD objects will not be written! If the subdirectory exists, several files will exist after the simulation is complete - they are in a format that is readable by libROM.
Expected screen output:
HyPar - Parallel (MPI) version with 16 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 2
No. of variables : 4
Domain size : 128 128
Processes along each dimension : 4 4
Exact solution domain size : 128 128
No. of ghosts pts : 3
No. of iter. : 50000
Restart iteration : 0
Time integration scheme : rk (44)
Spatial discretization scheme (hyperbolic) : upw5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : components
Spatial discretization type (parabolic ) : nonconservative-2stage
Spatial discretization scheme (parabolic ) : 4
Time Step : 5.000000E-03
Check for conservation : no
Screen output iterations : 1000
File output iterations : 10000
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : yes
Physical model : navierstokes2d
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 1.0158100316200525E+00
1: -1.7347234759768071E-17
2: -8.6736173798840355E-19
3: 1.8148063242358203E+00
Reading boundary conditions from boundary.inp.
Boundary noslip-wall: Along dimension 0 and face +1
Boundary noslip-wall: Along dimension 0 and face -1
Boundary noslip-wall: Along dimension 1 and face +1
Boundary noslip-wall: Along dimension 1 and face -1
4 boundary condition(s) read.
Initializing solvers.
Initializing physics. Model = "navierstokes2d"
Reading physical model inputs from file "physics.inp".
Setting up time integration.
Setting up libROM interface.
libROM inputs and parameters:
reduced model dimensionality: 16
sampling frequency: 25
mode: train
type: DMD
save to file: true
local vector size: 4096
libROM DMD inputs:
number of samples per window: 400
directory name for DMD onjects: DMD
Solving in time (from 0 to 50000 iterations)
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=0.000000 (total: 1).
iter= 1000 t=5.000E+00 CFL=7.012E-01 norm=2.9995E-03 wctime: 1.1E-02 (s)
iter= 2000 t=1.000E+01 CFL=7.023E-01 norm=2.1289E-03 wctime: 1.1E-02 (s)
iter= 3000 t=1.500E+01 CFL=7.092E-01 norm=2.1891E-03 wctime: 1.1E-02 (s)
iter= 4000 t=2.000E+01 CFL=7.039E-01 norm=1.7303E-03 wctime: 1.1E-02 (s)
iter= 5000 t=2.500E+01 CFL=7.004E-01 norm=1.3120E-03 wctime: 1.1E-02 (s)
iter= 6000 t=3.000E+01 CFL=6.968E-01 norm=1.1986E-03 wctime: 1.1E-02 (s)
iter= 7000 t=3.500E+01 CFL=6.961E-01 norm=1.2731E-03 wctime: 1.1E-02 (s)
iter= 8000 t=4.000E+01 CFL=6.955E-01 norm=9.5996E-04 wctime: 1.1E-02 (s)
iter= 9000 t=4.500E+01 CFL=6.956E-01 norm=8.6183E-04 wctime: 1.1E-02 (s)
iter= 10000 t=5.000E+01 CFL=6.964E-01 norm=8.1264E-04 wctime: 1.1E-02 (s)
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=50.000000 (total: 2).
iter= 11000 t=5.500E+01 CFL=6.987E-01 norm=7.8505E-04 wctime: 1.1E-02 (s)
iter= 12000 t=6.000E+01 CFL=6.989E-01 norm=6.2630E-04 wctime: 1.1E-02 (s)
iter= 13000 t=6.500E+01 CFL=6.927E-01 norm=5.2062E-04 wctime: 1.1E-02 (s)
iter= 14000 t=7.000E+01 CFL=6.943E-01 norm=5.6029E-04 wctime: 1.1E-02 (s)
iter= 15000 t=7.500E+01 CFL=6.929E-01 norm=4.6944E-04 wctime: 1.1E-02 (s)
iter= 16000 t=8.000E+01 CFL=6.949E-01 norm=3.8959E-04 wctime: 1.1E-02 (s)
iter= 17000 t=8.500E+01 CFL=6.947E-01 norm=3.1033E-04 wctime: 1.1E-02 (s)
iter= 18000 t=9.000E+01 CFL=6.957E-01 norm=3.7533E-04 wctime: 1.1E-02 (s)
iter= 19000 t=9.500E+01 CFL=6.951E-01 norm=3.0507E-04 wctime: 1.1E-02 (s)
iter= 20000 t=1.000E+02 CFL=6.932E-01 norm=2.4821E-04 wctime: 1.1E-02 (s)
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=100.000000 (total: 3).
iter= 21000 t=1.050E+02 CFL=6.934E-01 norm=2.7306E-04 wctime: 1.1E-02 (s)
iter= 22000 t=1.100E+02 CFL=6.926E-01 norm=2.4066E-04 wctime: 1.1E-02 (s)
iter= 23000 t=1.150E+02 CFL=6.931E-01 norm=2.1484E-04 wctime: 1.1E-02 (s)
iter= 24000 t=1.200E+02 CFL=6.933E-01 norm=1.7314E-04 wctime: 1.1E-02 (s)
iter= 25000 t=1.250E+02 CFL=6.943E-01 norm=1.7899E-04 wctime: 1.1E-02 (s)
iter= 26000 t=1.300E+02 CFL=6.944E-01 norm=1.6131E-04 wctime: 1.1E-02 (s)
iter= 27000 t=1.350E+02 CFL=6.941E-01 norm=1.4382E-04 wctime: 1.1E-02 (s)
iter= 28000 t=1.400E+02 CFL=6.933E-01 norm=1.3690E-04 wctime: 1.1E-02 (s)
iter= 29000 t=1.450E+02 CFL=6.934E-01 norm=1.2948E-04 wctime: 1.1E-02 (s)
iter= 30000 t=1.500E+02 CFL=6.930E-01 norm=1.3045E-04 wctime: 1.1E-02 (s)
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=150.000000 (total: 4).
iter= 31000 t=1.550E+02 CFL=6.937E-01 norm=9.0917E-05 wctime: 1.1E-02 (s)
iter= 32000 t=1.600E+02 CFL=6.943E-01 norm=8.2598E-05 wctime: 1.1E-02 (s)
iter= 33000 t=1.650E+02 CFL=6.944E-01 norm=9.3604E-05 wctime: 1.1E-02 (s)
iter= 34000 t=1.700E+02 CFL=6.941E-01 norm=8.2717E-05 wctime: 1.1E-02 (s)
iter= 35000 t=1.750E+02 CFL=6.937E-01 norm=6.0937E-05 wctime: 1.1E-02 (s)
iter= 36000 t=1.800E+02 CFL=6.935E-01 norm=6.9049E-05 wctime: 1.1E-02 (s)
iter= 37000 t=1.850E+02 CFL=6.933E-01 norm=8.1859E-05 wctime: 1.1E-02 (s)
iter= 38000 t=1.900E+02 CFL=6.937E-01 norm=6.5221E-05 wctime: 1.1E-02 (s)
iter= 39000 t=1.950E+02 CFL=6.939E-01 norm=5.3237E-05 wctime: 1.1E-02 (s)
iter= 40000 t=2.000E+02 CFL=6.942E-01 norm=5.6340E-05 wctime: 1.1E-02 (s)
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=200.000000 (total: 5).
iter= 41000 t=2.050E+02 CFL=6.941E-01 norm=5.5951E-05 wctime: 1.1E-02 (s)
iter= 42000 t=2.100E+02 CFL=6.941E-01 norm=4.3380E-05 wctime: 1.1E-02 (s)
iter= 43000 t=2.150E+02 CFL=6.938E-01 norm=4.1924E-05 wctime: 1.1E-02 (s)
iter= 44000 t=2.200E+02 CFL=6.938E-01 norm=5.1997E-05 wctime: 1.1E-02 (s)
iter= 45000 t=2.250E+02 CFL=6.938E-01 norm=4.4642E-05 wctime: 1.1E-02 (s)
iter= 46000 t=2.300E+02 CFL=6.941E-01 norm=2.8358E-05 wctime: 1.1E-02 (s)
iter= 47000 t=2.350E+02 CFL=6.941E-01 norm=2.9105E-05 wctime: 1.1E-02 (s)
iter= 48000 t=2.400E+02 CFL=6.942E-01 norm=3.3503E-05 wctime: 1.1E-02 (s)
iter= 49000 t=2.450E+02 CFL=6.940E-01 norm=2.4237E-05 wctime: 1.1E-02 (s)
iter= 50000 t=2.500E+02 CFL=6.939E-01 norm=1.8765E-05 wctime: 1.1E-02 (s)
Completed time integration (Final time: 250.000000), total wctime: 554.569953 (seconds).
Writing solution file op.bin.
libROM: Training ROM.
DMDROMObject::train() - training DMD object 0 with 401 samples.
Using 16 basis vectors out of 400.
DMDROMObject::train() - training DMD object 1 with 401 samples.
Using 16 basis vectors out of 400.
DMDROMObject::train() - training DMD object 2 with 401 samples.
Using 16 basis vectors out of 400.
DMDROMObject::train() - training DMD object 3 with 401 samples.
Using 16 basis vectors out of 400.
DMDROMObject::train() - training DMD object 4 with 400 samples.
Using 16 basis vectors out of 399.
libROM: total training wallclock time: 58.575120 (seconds).
libROM: Predicting solution at time 0.0000e+00 using ROM.
libROM: wallclock time: 0.053246 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 5.0000e+01 using ROM.
libROM: wallclock time: 0.052539 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 1.0000e+02 using ROM.
libROM: wallclock time: 0.051701 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 1.5000e+02 using ROM.
libROM: wallclock time: 0.051391 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 2.0000e+02 using ROM.
libROM: wallclock time: 0.051721 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 2.5000e+02 using ROM.
libROM: wallclock time: 0.051367 (seconds).
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom.bin.
libROM: total prediction/query wallclock time: 0.311965 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object with filename root DMD/dmdobj_0000.
Saving DMD object with filename root DMD/dmdobj_0001.
Saving DMD object with filename root DMD/dmdobj_0002.
Saving DMD object with filename root DMD/dmdobj_0003.
Saving DMD object with filename root DMD/dmdobj_0004.
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 8.5992357949730321E-03
L2 Norm : 8.6116162558021424E-03
Linfinity Norm : 9.9877095768199000E-03
Solver runtime (in seconds): 6.1712231799999995E+02
Total runtime (in seconds): 6.1738959499999999E+02
Deallocating arrays.
Finished.