Reduced Order Modeling: This example trains a DMD object and then predicts the solution using the DMD at the same times that the actual HyPar solution is written at.
Domain: The domain consists of a fine uniform grid around the cylinder defined by [-2,6] X [-2,2], and a stretched grid beyond this zone. Note: This is a 2D flow simulated using a 3-dimensional setup by taking the length of the domain along z to be very small and with only 3 grid points (the domain size along z must be smaller than the cylinder length).
Initial solution: \(\rho=1, u=0.1, v=w=0, p=1/\gamma\) everywhere in the domain.
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
{
const double GAMMA = 1.4;
int NI,NJ,NK,ndims;
char ip_file_type[50];
FILE *in;
strcpy(ip_file_type,"ascii");
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) {
printf("Error: Input file \"solver.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")){
while (strcmp(word, "end")){
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) {
fscanf(in,"%d",&ndims);
} else if (!strcmp(word, "size" )) {
fscanf(in,"%d",&NI);
fscanf(in,"%d",&NJ);
fscanf(in,"%d",&NK);
} else if (!strcmp(word, "ip_file_type")) {
fscanf(in,"%s",ip_file_type);
}
}
} else {
printf("Error: Illegal format in solver.inp. Crash and burn!\n");
return(0);
}
}
fclose(in);
if (ndims != 3) {
printf("Error: ndims is not 3 in solver.inp.\n");
return(0);
}
printf("Grid: %3d x %3d x %3d.\n",NI,NJ,NK);
double Lx = 8.0;
double Ly = 4.0;
double sf_x1 = 1.15;
double sf_x2 = 1.20;
double sf_y = 1.4;
int i,j,k;
double u = 0.1,
v = 0.0,
w = 0.0,
rho = 1.0,
P = 1.0/GAMMA;
double *x, *y, *z, *U;
double dx, dy, dz;
FILE *out;
x = (double*) calloc (NI , sizeof(double));
y = (double*) calloc (NJ , sizeof(double));
z = (double*) calloc (NK , sizeof(double));
U = (double*) calloc (5*NI*NJ*NK, sizeof(double));
x[NI/8] = -Lx/4;
x[7*NI/8] = 3*Lx/4;
dx = Lx / ((double)(6*NI/8));
for (i = NI/8 ; i < 7*NI/8; i++) x[i] = x[NI/8] + dx * (i-NI/8);
for (i = 7*NI/8; i < NI ; i++) x[i] = x[i-1] + sf_x2 * (x[i-1] - x[i-2]);
for (i = NI/8-1; i >= 0 ; i--) x[i] = x[i+1] - sf_x1 * (x[i+2] - x[i+1]);
y[NJ/8] = -Ly/2;
y[7*NJ/8] = Ly/2;
dy = Ly / ((double)(6*NJ/8));
for (j = NJ/8 ; j < 7*NJ/8; j++) y[j] = y[NJ/8] + dy * (j-NJ/8);
for (j = 7*NJ/8; j < NJ ; j++) y[j] = y[j-1] + sf_y * (y[j-1] - y[j-2]);
for (j = NJ/8-1; j >= 0 ; j--) y[j] = y[j+1] - sf_y * (y[j+2] - y[j+1]);
dz = 0.5 * (dx +dy);
for (k = 0; k < NK; k++) z[k] = (k-NK/2) * dz;
for (i = 0; i < NI; i++){
for (j = 0; j < NJ; j++){
for (k = 0; k < NK; k++){
int p = i + NI*j + NI*NJ*k;
U[5*p+0] = rho;
U[5*p+1] = rho*u;
U[5*p+2] = rho*v;
U[5*p+3] = rho*w;
U[5*p+4] = P/(GAMMA-1.0) + 0.5*rho*(u*u+v*v+w*w);
}
}
}
if (!strcmp(ip_file_type,"ascii")) {
printf("Writing ASCII initial solution file initial.inp\n");
out = fopen("initial.inp","w");
for (i = 0; i < NI; i++) fprintf(out,"%1.16E ",x[i]);
fprintf(out,"\n");
for (j = 0; j < NJ; j++) fprintf(out,"%1.16E ",y[j]);
fprintf(out,"\n");
for (k = 0; k < NK; k++) fprintf(out,"%1.16E ",z[k]);
fprintf(out,"\n");
for (k = 0; k < NK; k++) {
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = i + NK*j + NI*NJ*k;
fprintf(out,"%1.16E ",U[5*p+0]);
}
}
}
fprintf(out,"\n");
for (k = 0; k < NK; k++) {
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = i + NK*j + NI*NJ*k;
fprintf(out,"%1.16E ",U[5*p+1]);
}
}
}
fprintf(out,"\n");
for (k = 0; k < NK; k++) {
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = i + NK*j + NI*NJ*k;
fprintf(out,"%1.16E ",U[5*p+2]);
}
}
}
fprintf(out,"\n");
for (k = 0; k < NK; k++) {
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = i + NK*j + NI*NJ*k;
fprintf(out,"%1.16E ",U[5*p+3]);
}
}
}
fprintf(out,"\n");
for (k = 0; k < NK; k++) {
for (j = 0; j < NJ; j++) {
for (i = 0; i < NI; i++) {
int p = i + NK*j + NI*NJ*k;
fprintf(out,"%1.16E ",U[5*p+4]);
}
}
}
fprintf(out,"\n");
fclose(out);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary initial solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x,sizeof(double),NI,out);
fwrite(y,sizeof(double),NJ,out);
fwrite(z,sizeof(double),NK,out);
fwrite(U,sizeof(double),5*NI*NJ*NK,out);
fclose(out);
}
free(x);
free(y);
free(z);
free(U);
return(0);
}
Please see the original example in the "Immersed Boundaries Examples" section for a full description of the output files and how to view them. The following only contains libROM-specific comments.
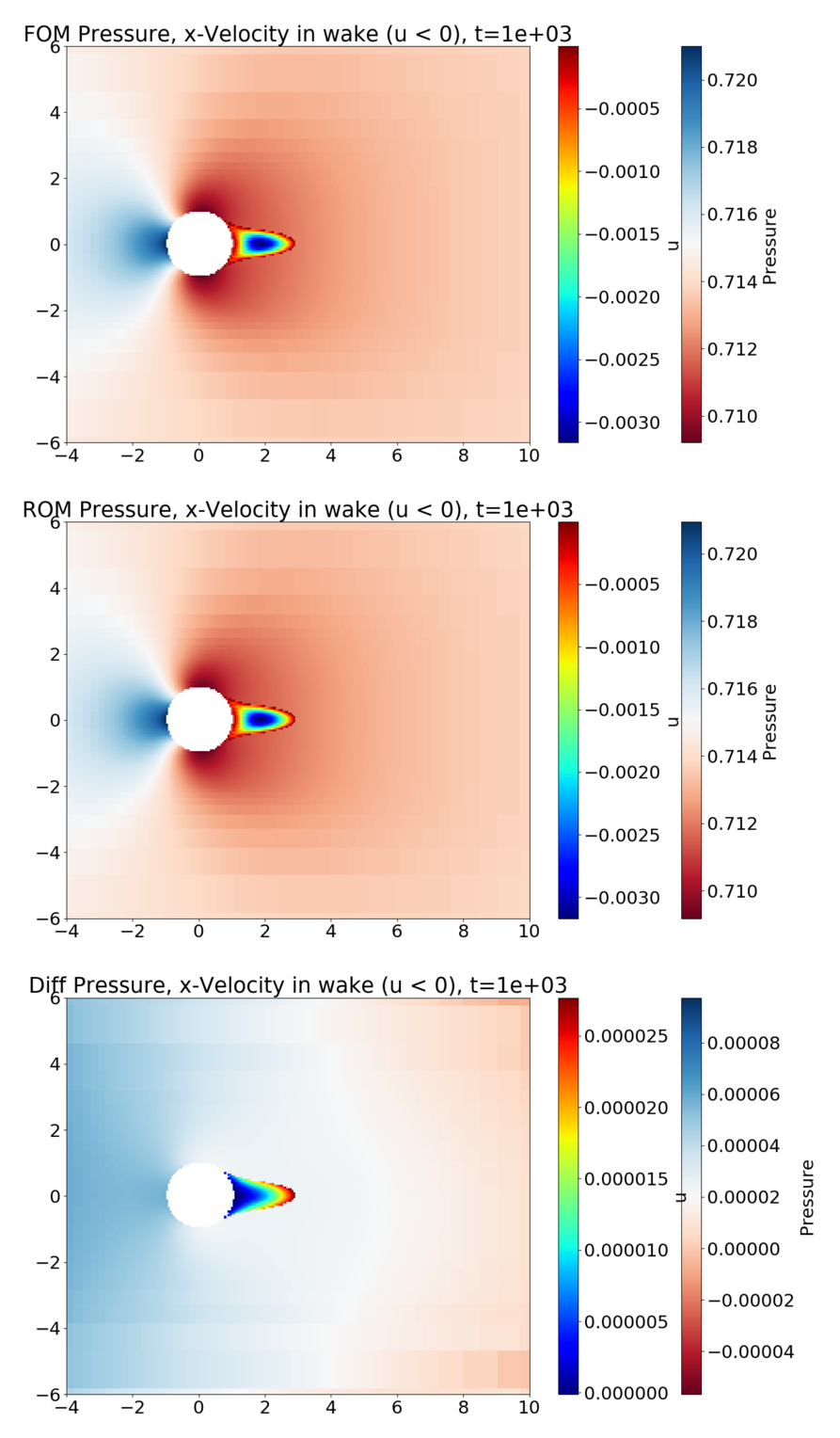
The following figure shows the flow at \(Re_D=20\). The pressure is plotted in the overall domain, and the wake is shown by plotting the x-velocity, where it is negative. FOM (full-order model) refers to the HyPar solution, ROM (reduced-order model) refers to the DMD solution, and Diff is the difference between the two.
HyPar - Parallel (MPI) version with 32 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 3
No. of variables : 5
Domain size : 200 96 3
Processes along each dimension : 8 4 1
Exact solution domain size : 200 96 3
No. of ghosts pts : 3
No. of iter. : 25000
Restart iteration : 0
Time integration scheme : rk (44)
Spatial discretization scheme (hyperbolic) : weno5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : components
Spatial discretization type (parabolic ) : nonconservative-2stage
Spatial discretization scheme (parabolic ) : 4
Time Step : 4.000000E-02
Check for conservation : no
Screen output iterations : 500
File output iterations : 2500
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : yes
Physical model : navierstokes3d
Immersed Body : cylinder.stl
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 2.5654236261034112E+02
1: 2.5654236261034061E+01
2: 0.0000000000000000E+00
3: 0.0000000000000000E+00
4: 4.5939407361723192E+02
Reading boundary conditions from boundary.inp.
Boundary subsonic-inflow: Along dimension 0 and face +1
Boundary subsonic-outflow: Along dimension 0 and face -1
Boundary subsonic-ambivalent: Along dimension 1 and face +1
Boundary subsonic-ambivalent: Along dimension 1 and face -1
Boundary periodic: Along dimension 2 and face +1
Boundary periodic: Along dimension 2 and face -1
6 boundary condition(s) read.
Immersed body read from cylinder.stl:
Number of facets: 628
Bounding box: [-1.0,1.0] X [-1.0,1.0] X [-5.0,5.0]
Number of grid points inside immersed body: 3456 ( 6.0%).
Number of immersed boundary points : 882 ( 1.5%).
Immersed body simulation mode : 2d (xy).
Initializing solvers.
Warning: File weno.inp not found. Using default parameters for WENO5/CRWENO5/HCWENO5 scheme.
Initializing physics. Model = "navierstokes3d"
Reading physical model inputs from file "physics.inp".
Writing solution file iblank.bin.
Setting up time integration.
Setting up libROM interface.
libROM inputs and parameters:
reduced model dimensionality: 16
sampling frequency: 10
mode: train
type: DMD
save to file: true
local vector size: 9000
libROM DMD inputs:
number of samples per window: 500
directory name for DMD onjects: DMD
Solving in time (from 0 to 25000 iterations)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=0.000000 (total: 1).
iter= 500 t=2.000E+01 CFL=8.522E-01 norm=5.8781E-05 wctime: 1.2E-01 (s)
iter= 1000 t=4.000E+01 CFL=8.455E-01 norm=7.1795E-05 wctime: 1.2E-01 (s)
iter= 1500 t=6.000E+01 CFL=8.447E-01 norm=1.8942E-05 wctime: 1.2E-01 (s)
iter= 2000 t=8.000E+01 CFL=8.433E-01 norm=1.2998E-05 wctime: 1.2E-01 (s)
iter= 2500 t=1.000E+02 CFL=8.416E-01 norm=8.9780E-06 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
iter= 3000 t=1.200E+02 CFL=8.407E-01 norm=7.9503E-06 wctime: 1.2E-01 (s)
iter= 3500 t=1.400E+02 CFL=8.406E-01 norm=4.2791E-06 wctime: 1.2E-01 (s)
iter= 4000 t=1.600E+02 CFL=8.401E-01 norm=4.8907E-06 wctime: 1.2E-01 (s)
iter= 4500 t=1.800E+02 CFL=8.402E-01 norm=3.2235E-06 wctime: 1.2E-01 (s)
iter= 5000 t=2.000E+02 CFL=8.396E-01 norm=3.0426E-06 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=200.000000 (total: 2).
iter= 5500 t=2.200E+02 CFL=8.393E-01 norm=4.3116E-06 wctime: 1.2E-01 (s)
iter= 6000 t=2.400E+02 CFL=8.393E-01 norm=2.1752E-06 wctime: 1.2E-01 (s)
iter= 6500 t=2.600E+02 CFL=8.391E-01 norm=1.6227E-06 wctime: 1.2E-01 (s)
iter= 7000 t=2.800E+02 CFL=8.391E-01 norm=1.4332E-06 wctime: 1.2E-01 (s)
iter= 7500 t=3.000E+02 CFL=8.389E-01 norm=1.8285E-06 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
iter= 8000 t=3.200E+02 CFL=8.388E-01 norm=1.4675E-06 wctime: 1.2E-01 (s)
iter= 8500 t=3.400E+02 CFL=8.387E-01 norm=1.0198E-06 wctime: 1.2E-01 (s)
iter= 9000 t=3.600E+02 CFL=8.388E-01 norm=1.2607E-06 wctime: 1.2E-01 (s)
iter= 9500 t=3.800E+02 CFL=8.385E-01 norm=1.2296E-06 wctime: 1.2E-01 (s)
iter= 10000 t=4.000E+02 CFL=8.386E-01 norm=9.3076E-07 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=400.000000 (total: 3).
iter= 10500 t=4.200E+02 CFL=8.384E-01 norm=1.0897E-06 wctime: 1.2E-01 (s)
iter= 11000 t=4.400E+02 CFL=8.385E-01 norm=1.7621E-06 wctime: 1.2E-01 (s)
iter= 11500 t=4.600E+02 CFL=8.384E-01 norm=1.3856E-06 wctime: 1.2E-01 (s)
iter= 12000 t=4.800E+02 CFL=8.384E-01 norm=7.9564E-07 wctime: 1.2E-01 (s)
iter= 12500 t=5.000E+02 CFL=8.383E-01 norm=7.0599E-07 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
iter= 13000 t=5.200E+02 CFL=8.384E-01 norm=7.9634E-07 wctime: 1.2E-01 (s)
iter= 13500 t=5.400E+02 CFL=8.382E-01 norm=6.0116E-07 wctime: 1.2E-01 (s)
iter= 14000 t=5.600E+02 CFL=8.383E-01 norm=5.7136E-07 wctime: 1.2E-01 (s)
iter= 14500 t=5.800E+02 CFL=8.382E-01 norm=9.7077E-07 wctime: 1.2E-01 (s)
iter= 15000 t=6.000E+02 CFL=8.381E-01 norm=1.1009E-06 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=600.000000 (total: 4).
iter= 15500 t=6.200E+02 CFL=8.381E-01 norm=6.4428E-07 wctime: 1.2E-01 (s)
iter= 16000 t=6.400E+02 CFL=8.381E-01 norm=3.9152E-07 wctime: 1.2E-01 (s)
iter= 16500 t=6.600E+02 CFL=8.380E-01 norm=5.6488E-07 wctime: 1.2E-01 (s)
iter= 17000 t=6.800E+02 CFL=8.381E-01 norm=5.1070E-07 wctime: 1.2E-01 (s)
iter= 17500 t=7.000E+02 CFL=8.380E-01 norm=3.0742E-07 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
iter= 18000 t=7.200E+02 CFL=8.380E-01 norm=5.8484E-07 wctime: 1.2E-01 (s)
iter= 18500 t=7.400E+02 CFL=8.380E-01 norm=7.9417E-07 wctime: 1.2E-01 (s)
iter= 19000 t=7.600E+02 CFL=8.380E-01 norm=6.2746E-07 wctime: 1.2E-01 (s)
iter= 19500 t=7.800E+02 CFL=8.379E-01 norm=1.9335E-07 wctime: 1.2E-01 (s)
iter= 20000 t=8.000E+02 CFL=8.380E-01 norm=4.3277E-07 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
DMDROMObject::takeSample() - creating new DMD object, t=800.000000 (total: 5).
iter= 20500 t=8.200E+02 CFL=8.379E-01 norm=4.9462E-07 wctime: 1.2E-01 (s)
iter= 21000 t=8.400E+02 CFL=8.379E-01 norm=2.1972E-07 wctime: 1.2E-01 (s)
iter= 21500 t=8.600E+02 CFL=8.379E-01 norm=3.2268E-07 wctime: 1.2E-01 (s)
iter= 22000 t=8.800E+02 CFL=8.379E-01 norm=5.8261E-07 wctime: 1.2E-01 (s)
iter= 22500 t=9.000E+02 CFL=8.379E-01 norm=5.2245E-07 wctime: 1.2E-01 (s)
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
iter= 23000 t=9.200E+02 CFL=8.378E-01 norm=2.4261E-07 wctime: 1.2E-01 (s)
iter= 23500 t=9.400E+02 CFL=8.378E-01 norm=2.9603E-07 wctime: 1.2E-01 (s)
iter= 24000 t=9.600E+02 CFL=8.378E-01 norm=4.5186E-07 wctime: 1.2E-01 (s)
iter= 24500 t=9.800E+02 CFL=8.378E-01 norm=3.1621E-07 wctime: 1.2E-01 (s)
iter= 25000 t=1.000E+03 CFL=8.378E-01 norm=1.5889E-07 wctime: 1.2E-01 (s)
Completed time integration (Final time: 1000.000000), total wctime: 3037.759859 (seconds).
Writing immersed body surface data file surface.dat.
Writing solution file op.bin.
libROM: Training ROM.
DMDROMObject::train() - training DMD object 0 with 501 samples.
Using 16 basis vectors out of 500.
DMDROMObject::train() - training DMD object 1 with 501 samples.
Using 16 basis vectors out of 500.
DMDROMObject::train() - training DMD object 2 with 501 samples.
Using 16 basis vectors out of 500.
DMDROMObject::train() - training DMD object 3 with 501 samples.
Using 16 basis vectors out of 500.
DMDROMObject::train() - training DMD object 4 with 500 samples.
Using 16 basis vectors out of 499.
libROM: total training wallclock time: 248.867239 (seconds).
libROM: Predicting solution at time 0.0000e+00 using ROM.
libROM: wallclock time: 0.124105 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 1.0000e+02 using ROM.
libROM: wallclock time: 0.122800 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 2.0000e+02 using ROM.
libROM: wallclock time: 0.118202 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 3.0000e+02 using ROM.
libROM: wallclock time: 0.122758 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 4.0000e+02 using ROM.
libROM: wallclock time: 0.117810 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 5.0000e+02 using ROM.
libROM: wallclock time: 0.118294 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 6.0000e+02 using ROM.
libROM: wallclock time: 0.115538 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 7.0000e+02 using ROM.
libROM: wallclock time: 0.115497 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 8.0000e+02 using ROM.
libROM: wallclock time: 0.120119 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 9.0000e+02 using ROM.
libROM: wallclock time: 0.120292 (seconds).
Writing solution file op_rom.bin.
libROM: Predicting solution at time 1.0000e+03 using ROM.
libROM: wallclock time: 0.120184 (seconds).
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom.bin.
libROM: total prediction/query wallclock time: 1.315599 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object with filename root DMD/dmdobj_0000.
Saving DMD object with filename root DMD/dmdobj_0001.
Saving DMD object with filename root DMD/dmdobj_0002.
Saving DMD object with filename root DMD/dmdobj_0003.
Saving DMD object with filename root DMD/dmdobj_0004.
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 1.1558687642990055E-04
L2 Norm : 1.1862964044372682E-04
Linfinity Norm : 5.6357008384349757E-04
Solver runtime (in seconds): 3.2943461360000001E+03
Total runtime (in seconds): 3.2948568040000000E+03
Deallocating arrays.
Finished.