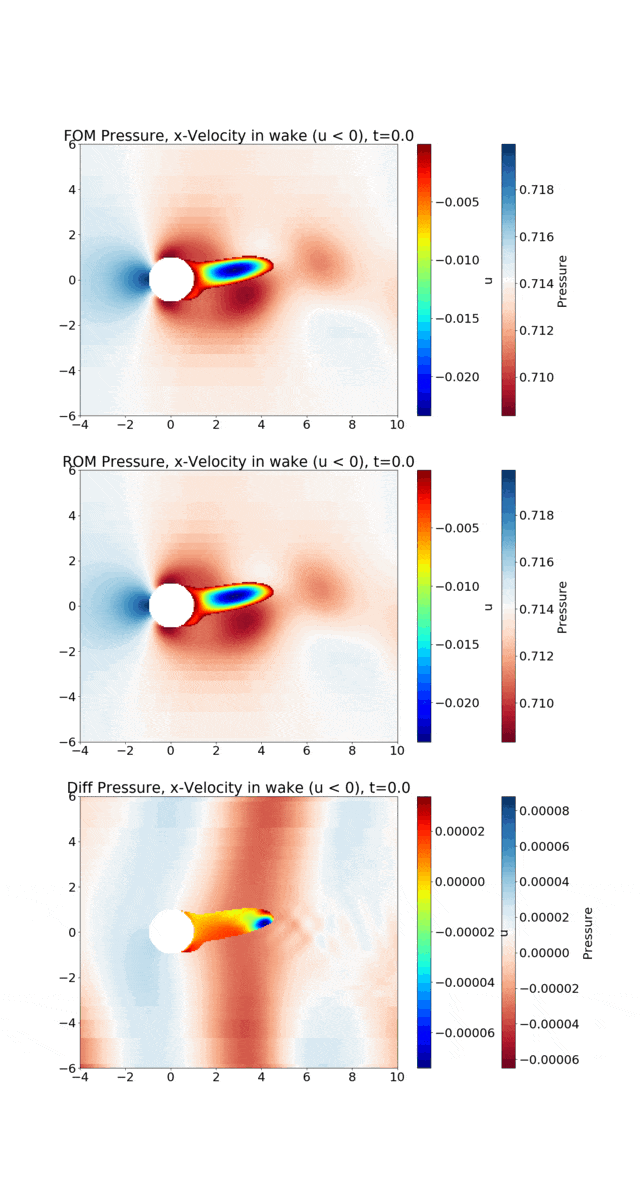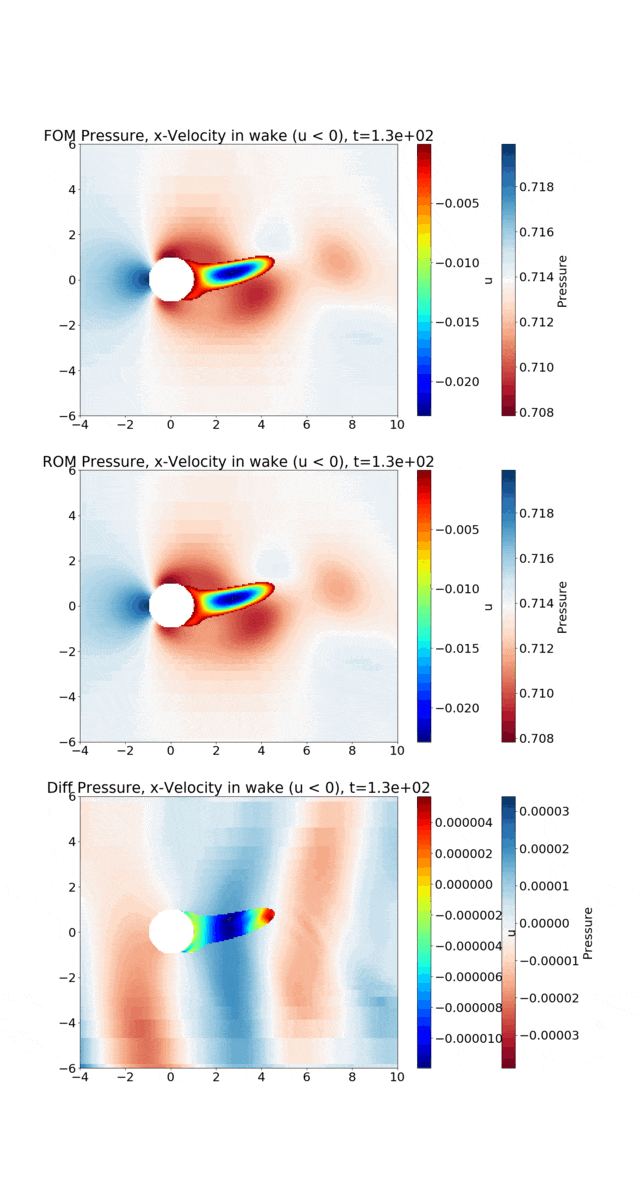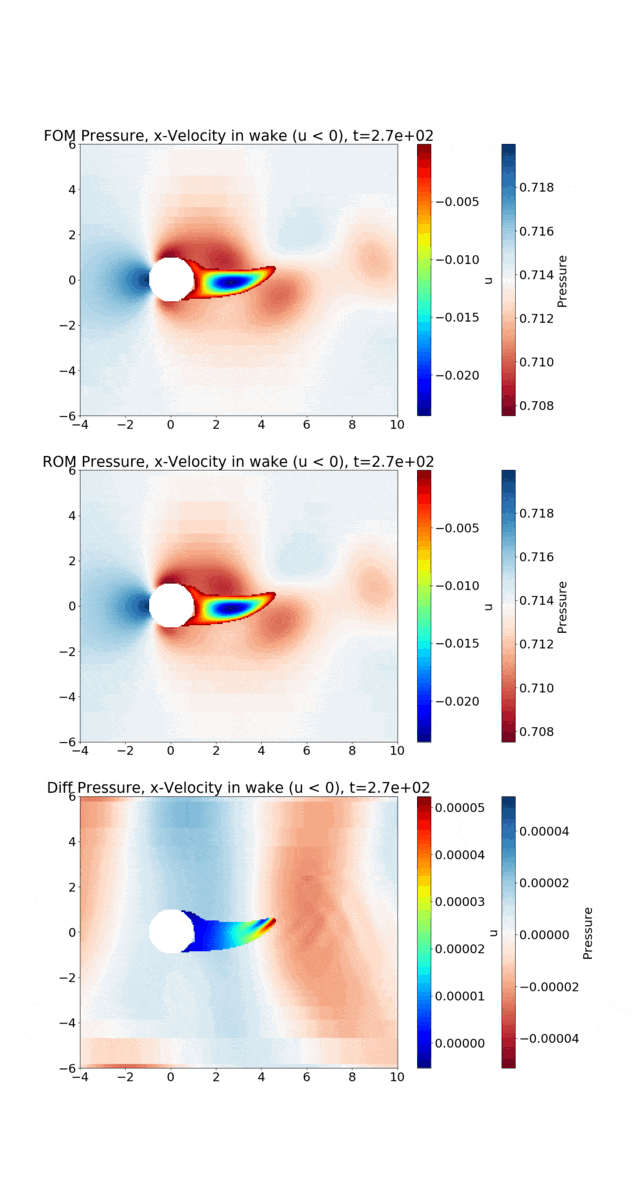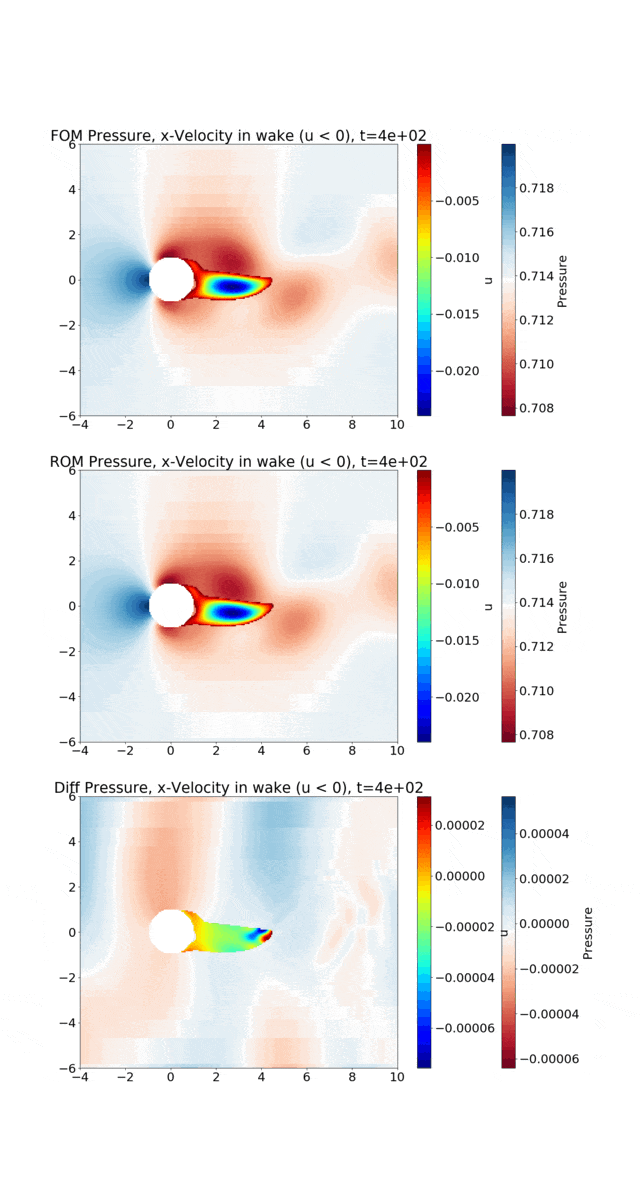
Reduced Order Modeling: This example trains a DMD object and then predicts the solution using the DMD at the same times that the actual HyPar solution is written at.
Domain: The domain consists of a fine uniform grid around the cylinder defined by [-4,12] X [-2,2], and a stretched grid beyond this zone. Note: This is a 2D flow simulated using a 3-dimensional setup by taking the length of the domain along z to be very small and with only 3 grid points (the domain size along z must be smaller than the cylinder length).
Initial solution: \(\rho=1, u=0.1, v=w=0, p=1/\gamma\) everywhere in the domain.
Please see the original example in the "Immersed Boundaries Examples" section for a full description of the output files and how to view them. The following only contains libROM-specific comments.
The following figure shows the vortex shedding. The pressure is plotted in the overall domain, and the wake is shown by plotting the x-velocity, where it is negative. FOM (full-order model) refers to the HyPar solution, ROM (reduced-order model) refers to the DMD solution, and Diff is the difference between the two.
HyPar - Parallel (MPI) version with 32 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 3
No. of variables : 5
Domain size : 400 96 3
Processes along each dimension : 8 4 1
Exact solution domain size : 400 96 3
No. of ghosts pts : 3
No. of iter. : 10000
Restart iteration : 0
Time integration scheme : rk (44)
Spatial discretization scheme (hyperbolic) : weno5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : components
Spatial discretization type (parabolic ) : nonconservative-2stage
Spatial discretization scheme (parabolic ) : 4
Time Step : 4.000000E-02
Check for conservation : no
Screen output iterations : 50
File output iterations : 250
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : no
Physical model : navierstokes3d
Immersed Body : cylinder.stl
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 2.9233368196521371E+02
1: 2.8593271918010952E+01
2: 9.8236287231872887E-03
3: -4.7954769269532492E-12
4: 5.2362439317196799E+02
Reading boundary conditions from boundary.inp.
Boundary subsonic-inflow: Along dimension 0 and face +1
Boundary subsonic-outflow: Along dimension 0 and face -1
Boundary subsonic-ambivalent: Along dimension 1 and face +1
Boundary subsonic-ambivalent: Along dimension 1 and face -1
Boundary periodic: Along dimension 2 and face +1
Boundary periodic: Along dimension 2 and face -1
6 boundary condition(s) read.
Immersed body read from cylinder.stl:
Number of facets: 628
Bounding box: [-1.0,1.0] X [-1.0,1.0] X [-5.0,5.0]
Number of grid points inside immersed body: 3264 ( 2.8%).
Number of immersed boundary points : 864 ( 0.8%).
Immersed body simulation mode : 2d (xy).
Initializing solvers.
Warning: File weno.inp not found. Using default parameters for WENO5/CRWENO5/HCWENO5 scheme.
Initializing physics. Model = "navierstokes3d"
Reading physical model inputs from file "physics.inp".
Writing solution file iblank_00000.bin.
Setting up time integration.
Setting up libROM interface.
libROM inputs and parameters:
reduced model dimensionality: 16
sampling frequency: 2
mode: train
type: DMD
save to file: true
local vector size: 18000
libROM DMD inputs:
number of samples per window: 1000
directory name for DMD onjects: DMD
Solving in time (from 0 to 10000 iterations)
Writing immersed body surface data file surface00000.dat.
Writing solution file op_00000.bin.
DMDROMObject::takeSample() - creating new DMD object, t=0.000000 (total: 1).
iter= 50 t=2.000E+00 CFL=8.485E-01 norm=6.2689E-05 wctime: 2.4E-01 (s)
iter= 100 t=4.000E+00 CFL=8.486E-01 norm=6.2661E-05 wctime: 2.5E-01 (s)
iter= 150 t=6.000E+00 CFL=8.486E-01 norm=6.2530E-05 wctime: 2.5E-01 (s)
iter= 200 t=8.000E+00 CFL=8.486E-01 norm=6.2243E-05 wctime: 2.5E-01 (s)
iter= 250 t=1.000E+01 CFL=8.487E-01 norm=6.1896E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00001.dat.
Writing solution file op_00001.bin.
iter= 300 t=1.200E+01 CFL=8.486E-01 norm=6.1456E-05 wctime: 2.5E-01 (s)
iter= 350 t=1.400E+01 CFL=8.486E-01 norm=6.0973E-05 wctime: 2.5E-01 (s)
iter= 400 t=1.600E+01 CFL=8.485E-01 norm=6.0543E-05 wctime: 2.5E-01 (s)
iter= 450 t=1.800E+01 CFL=8.484E-01 norm=6.0102E-05 wctime: 2.5E-01 (s)
iter= 500 t=2.000E+01 CFL=8.483E-01 norm=5.9813E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00002.dat.
Writing solution file op_00002.bin.
iter= 550 t=2.200E+01 CFL=8.483E-01 norm=5.9656E-05 wctime: 2.4E-01 (s)
iter= 600 t=2.400E+01 CFL=8.482E-01 norm=5.9581E-05 wctime: 2.5E-01 (s)
iter= 650 t=2.600E+01 CFL=8.484E-01 norm=5.9580E-05 wctime: 2.5E-01 (s)
iter= 700 t=2.800E+01 CFL=8.486E-01 norm=5.9716E-05 wctime: 2.5E-01 (s)
iter= 750 t=3.000E+01 CFL=8.488E-01 norm=5.9910E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00003.dat.
Writing solution file op_00003.bin.
iter= 800 t=3.200E+01 CFL=8.489E-01 norm=6.0099E-05 wctime: 2.5E-01 (s)
iter= 850 t=3.400E+01 CFL=8.489E-01 norm=6.0324E-05 wctime: 2.5E-01 (s)
iter= 900 t=3.600E+01 CFL=8.490E-01 norm=6.0541E-05 wctime: 2.5E-01 (s)
iter= 950 t=3.800E+01 CFL=8.490E-01 norm=6.0736E-05 wctime: 2.5E-01 (s)
iter= 1000 t=4.000E+01 CFL=8.491E-01 norm=6.0915E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00004.dat.
Writing solution file op_00004.bin.
iter= 1050 t=4.200E+01 CFL=8.490E-01 norm=6.1080E-05 wctime: 2.4E-01 (s)
iter= 1100 t=4.400E+01 CFL=8.490E-01 norm=6.1272E-05 wctime: 2.5E-01 (s)
iter= 1150 t=4.600E+01 CFL=8.489E-01 norm=6.1448E-05 wctime: 2.5E-01 (s)
iter= 1200 t=4.800E+01 CFL=8.488E-01 norm=6.1669E-05 wctime: 2.5E-01 (s)
iter= 1250 t=5.000E+01 CFL=8.486E-01 norm=6.1890E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00005.dat.
Writing solution file op_00005.bin.
iter= 1300 t=5.200E+01 CFL=8.484E-01 norm=6.2104E-05 wctime: 2.5E-01 (s)
iter= 1350 t=5.400E+01 CFL=8.484E-01 norm=6.2298E-05 wctime: 2.5E-01 (s)
iter= 1400 t=5.600E+01 CFL=8.485E-01 norm=6.2487E-05 wctime: 2.4E-01 (s)
iter= 1450 t=5.800E+01 CFL=8.486E-01 norm=6.2644E-05 wctime: 2.5E-01 (s)
iter= 1500 t=6.000E+01 CFL=8.486E-01 norm=6.2726E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00006.dat.
Writing solution file op_00006.bin.
iter= 1550 t=6.200E+01 CFL=8.486E-01 norm=6.2772E-05 wctime: 2.5E-01 (s)
iter= 1600 t=6.400E+01 CFL=8.487E-01 norm=6.2721E-05 wctime: 2.5E-01 (s)
iter= 1650 t=6.600E+01 CFL=8.487E-01 norm=6.2577E-05 wctime: 2.5E-01 (s)
iter= 1700 t=6.800E+01 CFL=8.487E-01 norm=6.2262E-05 wctime: 2.5E-01 (s)
iter= 1750 t=7.000E+01 CFL=8.486E-01 norm=6.1908E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00007.dat.
Writing solution file op_00007.bin.
iter= 1800 t=7.200E+01 CFL=8.486E-01 norm=6.1450E-05 wctime: 2.4E-01 (s)
iter= 1850 t=7.400E+01 CFL=8.486E-01 norm=6.1002E-05 wctime: 2.5E-01 (s)
iter= 1900 t=7.600E+01 CFL=8.485E-01 norm=6.0564E-05 wctime: 2.5E-01 (s)
iter= 1950 t=7.800E+01 CFL=8.485E-01 norm=6.0154E-05 wctime: 2.5E-01 (s)
iter= 2000 t=8.000E+01 CFL=8.484E-01 norm=5.9927E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00008.dat.
Writing solution file op_00008.bin.
DMDROMObject::takeSample() - creating new DMD object, t=80.000000 (total: 2).
iter= 2050 t=8.200E+01 CFL=8.484E-01 norm=5.9813E-05 wctime: 2.5E-01 (s)
iter= 2100 t=8.400E+01 CFL=8.484E-01 norm=5.9767E-05 wctime: 2.5E-01 (s)
iter= 2150 t=8.600E+01 CFL=8.486E-01 norm=5.9814E-05 wctime: 2.4E-01 (s)
iter= 2200 t=8.800E+01 CFL=8.488E-01 norm=5.9978E-05 wctime: 2.4E-01 (s)
iter= 2250 t=9.000E+01 CFL=8.489E-01 norm=6.0177E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00009.dat.
Writing solution file op_00009.bin.
iter= 2300 t=9.200E+01 CFL=8.490E-01 norm=6.0389E-05 wctime: 2.5E-01 (s)
iter= 2350 t=9.400E+01 CFL=8.490E-01 norm=6.0628E-05 wctime: 2.5E-01 (s)
iter= 2400 t=9.600E+01 CFL=8.491E-01 norm=6.0850E-05 wctime: 2.5E-01 (s)
iter= 2450 t=9.800E+01 CFL=8.491E-01 norm=6.1058E-05 wctime: 2.4E-01 (s)
iter= 2500 t=1.000E+02 CFL=8.491E-01 norm=6.1223E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00010.dat.
Writing solution file op_00010.bin.
iter= 2550 t=1.020E+02 CFL=8.491E-01 norm=6.1401E-05 wctime: 2.5E-01 (s)
iter= 2600 t=1.040E+02 CFL=8.490E-01 norm=6.1589E-05 wctime: 2.4E-01 (s)
iter= 2650 t=1.060E+02 CFL=8.489E-01 norm=6.1774E-05 wctime: 2.4E-01 (s)
iter= 2700 t=1.080E+02 CFL=8.487E-01 norm=6.1970E-05 wctime: 2.4E-01 (s)
iter= 2750 t=1.100E+02 CFL=8.485E-01 norm=6.2181E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00011.dat.
Writing solution file op_00011.bin.
iter= 2800 t=1.120E+02 CFL=8.484E-01 norm=6.2382E-05 wctime: 2.5E-01 (s)
iter= 2850 t=1.140E+02 CFL=8.484E-01 norm=6.2579E-05 wctime: 2.5E-01 (s)
iter= 2900 t=1.160E+02 CFL=8.485E-01 norm=6.2771E-05 wctime: 2.5E-01 (s)
iter= 2950 t=1.180E+02 CFL=8.486E-01 norm=6.2892E-05 wctime: 2.5E-01 (s)
iter= 3000 t=1.200E+02 CFL=8.486E-01 norm=6.2958E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00012.dat.
Writing solution file op_00012.bin.
iter= 3050 t=1.220E+02 CFL=8.486E-01 norm=6.3004E-05 wctime: 2.5E-01 (s)
iter= 3100 t=1.240E+02 CFL=8.487E-01 norm=6.2937E-05 wctime: 2.5E-01 (s)
iter= 3150 t=1.260E+02 CFL=8.487E-01 norm=6.2748E-05 wctime: 2.5E-01 (s)
iter= 3200 t=1.280E+02 CFL=8.487E-01 norm=6.2400E-05 wctime: 2.5E-01 (s)
iter= 3250 t=1.300E+02 CFL=8.487E-01 norm=6.2034E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00013.dat.
Writing solution file op_00013.bin.
iter= 3300 t=1.320E+02 CFL=8.487E-01 norm=6.1534E-05 wctime: 2.5E-01 (s)
iter= 3350 t=1.340E+02 CFL=8.486E-01 norm=6.1073E-05 wctime: 2.5E-01 (s)
iter= 3400 t=1.360E+02 CFL=8.485E-01 norm=6.0609E-05 wctime: 2.5E-01 (s)
iter= 3450 t=1.380E+02 CFL=8.484E-01 norm=6.0232E-05 wctime: 2.4E-01 (s)
iter= 3500 t=1.400E+02 CFL=8.484E-01 norm=6.0017E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00014.dat.
Writing solution file op_00014.bin.
iter= 3550 t=1.420E+02 CFL=8.483E-01 norm=5.9886E-05 wctime: 2.5E-01 (s)
iter= 3600 t=1.440E+02 CFL=8.485E-01 norm=5.9852E-05 wctime: 2.5E-01 (s)
iter= 3650 t=1.460E+02 CFL=8.487E-01 norm=5.9925E-05 wctime: 2.5E-01 (s)
iter= 3700 t=1.480E+02 CFL=8.489E-01 norm=6.0075E-05 wctime: 2.5E-01 (s)
iter= 3750 t=1.500E+02 CFL=8.490E-01 norm=6.0253E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00015.dat.
Writing solution file op_00015.bin.
iter= 3800 t=1.520E+02 CFL=8.490E-01 norm=6.0469E-05 wctime: 2.5E-01 (s)
iter= 3850 t=1.540E+02 CFL=8.491E-01 norm=6.0715E-05 wctime: 2.5E-01 (s)
iter= 3900 t=1.560E+02 CFL=8.491E-01 norm=6.0932E-05 wctime: 2.5E-01 (s)
iter= 3950 t=1.580E+02 CFL=8.492E-01 norm=6.1130E-05 wctime: 2.5E-01 (s)
iter= 4000 t=1.600E+02 CFL=8.491E-01 norm=6.1276E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00016.dat.
Writing solution file op_00016.bin.
DMDROMObject::takeSample() - creating new DMD object, t=160.000000 (total: 3).
iter= 4050 t=1.620E+02 CFL=8.491E-01 norm=6.1448E-05 wctime: 2.5E-01 (s)
iter= 4100 t=1.640E+02 CFL=8.490E-01 norm=6.1628E-05 wctime: 2.5E-01 (s)
iter= 4150 t=1.660E+02 CFL=8.489E-01 norm=6.1825E-05 wctime: 2.5E-01 (s)
iter= 4200 t=1.680E+02 CFL=8.487E-01 norm=6.2024E-05 wctime: 2.5E-01 (s)
iter= 4250 t=1.700E+02 CFL=8.485E-01 norm=6.2230E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00017.dat.
Writing solution file op_00017.bin.
iter= 4300 t=1.720E+02 CFL=8.484E-01 norm=6.2415E-05 wctime: 2.5E-01 (s)
iter= 4350 t=1.740E+02 CFL=8.485E-01 norm=6.2617E-05 wctime: 2.5E-01 (s)
iter= 4400 t=1.760E+02 CFL=8.486E-01 norm=6.2797E-05 wctime: 2.4E-01 (s)
iter= 4450 t=1.780E+02 CFL=8.486E-01 norm=6.2891E-05 wctime: 2.5E-01 (s)
iter= 4500 t=1.800E+02 CFL=8.487E-01 norm=6.2958E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00018.dat.
Writing solution file op_00018.bin.
iter= 4550 t=1.820E+02 CFL=8.487E-01 norm=6.2963E-05 wctime: 2.5E-01 (s)
iter= 4600 t=1.840E+02 CFL=8.488E-01 norm=6.2866E-05 wctime: 2.5E-01 (s)
iter= 4650 t=1.860E+02 CFL=8.487E-01 norm=6.2637E-05 wctime: 2.4E-01 (s)
iter= 4700 t=1.880E+02 CFL=8.487E-01 norm=6.2302E-05 wctime: 2.4E-01 (s)
iter= 4750 t=1.900E+02 CFL=8.487E-01 norm=6.1897E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00019.dat.
Writing solution file op_00019.bin.
iter= 4800 t=1.920E+02 CFL=8.486E-01 norm=6.1409E-05 wctime: 2.5E-01 (s)
iter= 4850 t=1.940E+02 CFL=8.486E-01 norm=6.0986E-05 wctime: 2.5E-01 (s)
iter= 4900 t=1.960E+02 CFL=8.485E-01 norm=6.0557E-05 wctime: 2.4E-01 (s)
iter= 4950 t=1.980E+02 CFL=8.485E-01 norm=6.0232E-05 wctime: 2.5E-01 (s)
iter= 5000 t=2.000E+02 CFL=8.484E-01 norm=6.0050E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00020.dat.
Writing solution file op_00020.bin.
iter= 5050 t=2.020E+02 CFL=8.483E-01 norm=5.9962E-05 wctime: 2.4E-01 (s)
iter= 5100 t=2.040E+02 CFL=8.486E-01 norm=5.9987E-05 wctime: 2.5E-01 (s)
iter= 5150 t=2.060E+02 CFL=8.488E-01 norm=6.0092E-05 wctime: 2.5E-01 (s)
iter= 5200 t=2.080E+02 CFL=8.489E-01 norm=6.0260E-05 wctime: 2.5E-01 (s)
iter= 5250 t=2.100E+02 CFL=8.490E-01 norm=6.0449E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00021.dat.
Writing solution file op_00021.bin.
iter= 5300 t=2.120E+02 CFL=8.491E-01 norm=6.0672E-05 wctime: 2.5E-01 (s)
iter= 5350 t=2.140E+02 CFL=8.492E-01 norm=6.0920E-05 wctime: 2.4E-01 (s)
iter= 5400 t=2.160E+02 CFL=8.492E-01 norm=6.1150E-05 wctime: 2.4E-01 (s)
iter= 5450 t=2.180E+02 CFL=8.492E-01 norm=6.1354E-05 wctime: 2.4E-01 (s)
iter= 5500 t=2.200E+02 CFL=8.491E-01 norm=6.1509E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00022.dat.
Writing solution file op_00022.bin.
iter= 5550 t=2.220E+02 CFL=8.491E-01 norm=6.1693E-05 wctime: 2.4E-01 (s)
iter= 5600 t=2.240E+02 CFL=8.490E-01 norm=6.1865E-05 wctime: 2.5E-01 (s)
iter= 5650 t=2.260E+02 CFL=8.489E-01 norm=6.2062E-05 wctime: 2.5E-01 (s)
iter= 5700 t=2.280E+02 CFL=8.487E-01 norm=6.2269E-05 wctime: 2.5E-01 (s)
iter= 5750 t=2.300E+02 CFL=8.485E-01 norm=6.2474E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00023.dat.
Writing solution file op_00023.bin.
iter= 5800 t=2.320E+02 CFL=8.485E-01 norm=6.2655E-05 wctime: 2.5E-01 (s)
iter= 5850 t=2.340E+02 CFL=8.485E-01 norm=6.2846E-05 wctime: 2.5E-01 (s)
iter= 5900 t=2.360E+02 CFL=8.486E-01 norm=6.3002E-05 wctime: 2.5E-01 (s)
iter= 5950 t=2.380E+02 CFL=8.487E-01 norm=6.3078E-05 wctime: 2.5E-01 (s)
iter= 6000 t=2.400E+02 CFL=8.487E-01 norm=6.3127E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00024.dat.
Writing solution file op_00024.bin.
DMDROMObject::takeSample() - creating new DMD object, t=240.000000 (total: 4).
iter= 6050 t=2.420E+02 CFL=8.487E-01 norm=6.3091E-05 wctime: 2.5E-01 (s)
iter= 6100 t=2.440E+02 CFL=8.487E-01 norm=6.2975E-05 wctime: 2.5E-01 (s)
iter= 6150 t=2.460E+02 CFL=8.488E-01 norm=6.2678E-05 wctime: 2.5E-01 (s)
iter= 6200 t=2.480E+02 CFL=8.488E-01 norm=6.2328E-05 wctime: 2.4E-01 (s)
iter= 6250 t=2.500E+02 CFL=8.487E-01 norm=6.1872E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00025.dat.
Writing solution file op_00025.bin.
iter= 6300 t=2.520E+02 CFL=8.487E-01 norm=6.1387E-05 wctime: 2.4E-01 (s)
iter= 6350 t=2.540E+02 CFL=8.486E-01 norm=6.0961E-05 wctime: 2.5E-01 (s)
iter= 6400 t=2.560E+02 CFL=8.485E-01 norm=6.0524E-05 wctime: 2.4E-01 (s)
iter= 6450 t=2.580E+02 CFL=8.484E-01 norm=6.0234E-05 wctime: 2.4E-01 (s)
iter= 6500 t=2.600E+02 CFL=8.483E-01 norm=6.0063E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00026.dat.
Writing solution file op_00026.bin.
iter= 6550 t=2.620E+02 CFL=8.484E-01 norm=5.9993E-05 wctime: 2.4E-01 (s)
iter= 6600 t=2.640E+02 CFL=8.486E-01 norm=6.0026E-05 wctime: 2.4E-01 (s)
iter= 6650 t=2.660E+02 CFL=8.488E-01 norm=6.0125E-05 wctime: 2.4E-01 (s)
iter= 6700 t=2.680E+02 CFL=8.490E-01 norm=6.0274E-05 wctime: 2.4E-01 (s)
iter= 6750 t=2.700E+02 CFL=8.491E-01 norm=6.0465E-05 wctime: 2.4E-01 (s)
Writing immersed body surface data file surface00027.dat.
Writing solution file op_00027.bin.
iter= 6800 t=2.720E+02 CFL=8.491E-01 norm=6.0706E-05 wctime: 2.4E-01 (s)
iter= 6850 t=2.740E+02 CFL=8.492E-01 norm=6.0943E-05 wctime: 2.5E-01 (s)
iter= 6900 t=2.760E+02 CFL=8.492E-01 norm=6.1150E-05 wctime: 2.5E-01 (s)
iter= 6950 t=2.780E+02 CFL=8.492E-01 norm=6.1338E-05 wctime: 2.5E-01 (s)
iter= 7000 t=2.800E+02 CFL=8.492E-01 norm=6.1499E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00028.dat.
Writing solution file op_00028.bin.
iter= 7050 t=2.820E+02 CFL=8.491E-01 norm=6.1682E-05 wctime: 2.4E-01 (s)
iter= 7100 t=2.840E+02 CFL=8.490E-01 norm=6.1870E-05 wctime: 2.5E-01 (s)
iter= 7150 t=2.860E+02 CFL=8.488E-01 norm=6.2073E-05 wctime: 2.5E-01 (s)
iter= 7200 t=2.880E+02 CFL=8.486E-01 norm=6.2266E-05 wctime: 2.4E-01 (s)
iter= 7250 t=2.900E+02 CFL=8.484E-01 norm=6.2455E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00029.dat.
Writing solution file op_00029.bin.
iter= 7300 t=2.920E+02 CFL=8.485E-01 norm=6.2654E-05 wctime: 2.5E-01 (s)
iter= 7350 t=2.940E+02 CFL=8.486E-01 norm=6.2862E-05 wctime: 2.5E-01 (s)
iter= 7400 t=2.960E+02 CFL=8.486E-01 norm=6.2971E-05 wctime: 2.5E-01 (s)
iter= 7450 t=2.980E+02 CFL=8.487E-01 norm=6.3026E-05 wctime: 2.4E-01 (s)
iter= 7500 t=3.000E+02 CFL=8.487E-01 norm=6.3058E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00030.dat.
Writing solution file op_00030.bin.
iter= 7550 t=3.020E+02 CFL=8.488E-01 norm=6.3002E-05 wctime: 2.5E-01 (s)
iter= 7600 t=3.040E+02 CFL=8.488E-01 norm=6.2842E-05 wctime: 2.5E-01 (s)
iter= 7650 t=3.060E+02 CFL=8.488E-01 norm=6.2509E-05 wctime: 2.5E-01 (s)
iter= 7700 t=3.080E+02 CFL=8.487E-01 norm=6.2137E-05 wctime: 2.5E-01 (s)
iter= 7750 t=3.100E+02 CFL=8.487E-01 norm=6.1661E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00031.dat.
Writing solution file op_00031.bin.
iter= 7800 t=3.120E+02 CFL=8.486E-01 norm=6.1223E-05 wctime: 2.5E-01 (s)
iter= 7850 t=3.140E+02 CFL=8.486E-01 norm=6.0814E-05 wctime: 2.5E-01 (s)
iter= 7900 t=3.160E+02 CFL=8.485E-01 norm=6.0434E-05 wctime: 2.5E-01 (s)
iter= 7950 t=3.180E+02 CFL=8.484E-01 norm=6.0208E-05 wctime: 2.5E-01 (s)
iter= 8000 t=3.200E+02 CFL=8.484E-01 norm=6.0083E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00032.dat.
Writing solution file op_00032.bin.
DMDROMObject::takeSample() - creating new DMD object, t=320.000000 (total: 5).
iter= 8050 t=3.220E+02 CFL=8.485E-01 norm=6.0069E-05 wctime: 2.5E-01 (s)
iter= 8100 t=3.240E+02 CFL=8.487E-01 norm=6.0140E-05 wctime: 2.5E-01 (s)
iter= 8150 t=3.260E+02 CFL=8.489E-01 norm=6.0286E-05 wctime: 2.5E-01 (s)
iter= 8200 t=3.280E+02 CFL=8.490E-01 norm=6.0462E-05 wctime: 2.4E-01 (s)
iter= 8250 t=3.300E+02 CFL=8.491E-01 norm=6.0654E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00033.dat.
Writing solution file op_00033.bin.
iter= 8300 t=3.320E+02 CFL=8.492E-01 norm=6.0878E-05 wctime: 2.4E-01 (s)
iter= 8350 t=3.340E+02 CFL=8.492E-01 norm=6.1124E-05 wctime: 2.4E-01 (s)
iter= 8400 t=3.360E+02 CFL=8.492E-01 norm=6.1342E-05 wctime: 2.4E-01 (s)
iter= 8450 t=3.380E+02 CFL=8.492E-01 norm=6.1513E-05 wctime: 2.4E-01 (s)
iter= 8500 t=3.400E+02 CFL=8.492E-01 norm=6.1691E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00034.dat.
Writing solution file op_00034.bin.
iter= 8550 t=3.420E+02 CFL=8.491E-01 norm=6.1873E-05 wctime: 2.5E-01 (s)
iter= 8600 t=3.440E+02 CFL=8.490E-01 norm=6.2068E-05 wctime: 2.5E-01 (s)
iter= 8650 t=3.460E+02 CFL=8.488E-01 norm=6.2277E-05 wctime: 2.5E-01 (s)
iter= 8700 t=3.480E+02 CFL=8.486E-01 norm=6.2496E-05 wctime: 2.5E-01 (s)
iter= 8750 t=3.500E+02 CFL=8.485E-01 norm=6.2689E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00035.dat.
Writing solution file op_00035.bin.
iter= 8800 t=3.520E+02 CFL=8.485E-01 norm=6.2868E-05 wctime: 2.4E-01 (s)
iter= 8850 t=3.540E+02 CFL=8.486E-01 norm=6.3045E-05 wctime: 2.4E-01 (s)
iter= 8900 t=3.560E+02 CFL=8.487E-01 norm=6.3141E-05 wctime: 2.5E-01 (s)
iter= 8950 t=3.580E+02 CFL=8.487E-01 norm=6.3183E-05 wctime: 2.5E-01 (s)
iter= 9000 t=3.600E+02 CFL=8.487E-01 norm=6.3180E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00036.dat.
Writing solution file op_00036.bin.
iter= 9050 t=3.620E+02 CFL=8.487E-01 norm=6.3096E-05 wctime: 2.5E-01 (s)
iter= 9100 t=3.640E+02 CFL=8.488E-01 norm=6.2862E-05 wctime: 2.5E-01 (s)
iter= 9150 t=3.660E+02 CFL=8.488E-01 norm=6.2497E-05 wctime: 2.5E-01 (s)
iter= 9200 t=3.680E+02 CFL=8.487E-01 norm=6.2085E-05 wctime: 2.5E-01 (s)
iter= 9250 t=3.700E+02 CFL=8.487E-01 norm=6.1596E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00037.dat.
Writing solution file op_00037.bin.
iter= 9300 t=3.720E+02 CFL=8.486E-01 norm=6.1158E-05 wctime: 2.5E-01 (s)
iter= 9350 t=3.740E+02 CFL=8.485E-01 norm=6.0715E-05 wctime: 2.5E-01 (s)
iter= 9400 t=3.760E+02 CFL=8.485E-01 norm=6.0374E-05 wctime: 2.5E-01 (s)
iter= 9450 t=3.780E+02 CFL=8.484E-01 norm=6.0178E-05 wctime: 2.4E-01 (s)
iter= 9500 t=3.800E+02 CFL=8.483E-01 norm=6.0074E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00038.dat.
Writing solution file op_00038.bin.
iter= 9550 t=3.820E+02 CFL=8.486E-01 norm=6.0074E-05 wctime: 2.5E-01 (s)
iter= 9600 t=3.840E+02 CFL=8.488E-01 norm=6.0152E-05 wctime: 2.4E-01 (s)
iter= 9650 t=3.860E+02 CFL=8.490E-01 norm=6.0292E-05 wctime: 2.5E-01 (s)
iter= 9700 t=3.880E+02 CFL=8.491E-01 norm=6.0468E-05 wctime: 2.5E-01 (s)
iter= 9750 t=3.900E+02 CFL=8.491E-01 norm=6.0681E-05 wctime: 2.5E-01 (s)
Writing immersed body surface data file surface00039.dat.
Writing solution file op_00039.bin.
iter= 9800 t=3.920E+02 CFL=8.492E-01 norm=6.0919E-05 wctime: 2.5E-01 (s)
iter= 9850 t=3.940E+02 CFL=8.492E-01 norm=6.1130E-05 wctime: 2.5E-01 (s)
iter= 9900 t=3.960E+02 CFL=8.492E-01 norm=6.1325E-05 wctime: 2.5E-01 (s)
iter= 9950 t=3.980E+02 CFL=8.492E-01 norm=6.1487E-05 wctime: 2.5E-01 (s)
iter= 10000 t=4.000E+02 CFL=8.491E-01 norm=6.1654E-05 wctime: 2.5E-01 (s)
Completed time integration (Final time: 400.000000), total wctime: 2413.348070 (seconds).
Writing immersed body surface data file surface00040.dat.
Writing solution file op_00040.bin.
libROM: Training ROM.
DMDROMObject::train() - training DMD object 0 with 1001 samples.
Using 16 basis vectors out of 1000.
DMDROMObject::train() - training DMD object 1 with 1001 samples.
Using 16 basis vectors out of 1000.
DMDROMObject::train() - training DMD object 2 with 1001 samples.
Using 16 basis vectors out of 1000.
DMDROMObject::train() - training DMD object 3 with 1001 samples.
Using 16 basis vectors out of 1000.
DMDROMObject::train() - training DMD object 4 with 1000 samples.
Using 16 basis vectors out of 999.
libROM: total training wallclock time: 2112.749892 (seconds).
libROM: Predicting solution at time 0.0000e+00 using ROM.
libROM: wallclock time: 0.233794 (seconds).
Writing solution file op_rom_00000.bin.
libROM: Predicting solution at time 1.0000e+01 using ROM.
libROM: wallclock time: 0.231340 (seconds).
Writing solution file op_rom_00001.bin.
libROM: Predicting solution at time 2.0000e+01 using ROM.
libROM: wallclock time: 0.234202 (seconds).
Writing solution file op_rom_00002.bin.
libROM: Predicting solution at time 3.0000e+01 using ROM.
libROM: wallclock time: 0.237055 (seconds).
Writing solution file op_rom_00003.bin.
libROM: Predicting solution at time 4.0000e+01 using ROM.
libROM: wallclock time: 0.236783 (seconds).
Writing solution file op_rom_00004.bin.
libROM: Predicting solution at time 5.0000e+01 using ROM.
libROM: wallclock time: 0.232317 (seconds).
Writing solution file op_rom_00005.bin.
libROM: Predicting solution at time 6.0000e+01 using ROM.
libROM: wallclock time: 0.237462 (seconds).
Writing solution file op_rom_00006.bin.
libROM: Predicting solution at time 7.0000e+01 using ROM.
libROM: wallclock time: 0.241502 (seconds).
Writing solution file op_rom_00007.bin.
libROM: Predicting solution at time 8.0000e+01 using ROM.
libROM: wallclock time: 0.246036 (seconds).
Writing solution file op_rom_00008.bin.
libROM: Predicting solution at time 9.0000e+01 using ROM.
libROM: wallclock time: 0.242724 (seconds).
Writing solution file op_rom_00009.bin.
libROM: Predicting solution at time 1.0000e+02 using ROM.
libROM: wallclock time: 0.244469 (seconds).
Writing solution file op_rom_00010.bin.
libROM: Predicting solution at time 1.1000e+02 using ROM.
libROM: wallclock time: 0.239162 (seconds).
Writing solution file op_rom_00011.bin.
libROM: Predicting solution at time 1.2000e+02 using ROM.
libROM: wallclock time: 0.239186 (seconds).
Writing solution file op_rom_00012.bin.
libROM: Predicting solution at time 1.3000e+02 using ROM.
libROM: wallclock time: 0.239854 (seconds).
Writing solution file op_rom_00013.bin.
libROM: Predicting solution at time 1.4000e+02 using ROM.
libROM: wallclock time: 0.240379 (seconds).
Writing solution file op_rom_00014.bin.
libROM: Predicting solution at time 1.5000e+02 using ROM.
libROM: wallclock time: 0.239745 (seconds).
Writing solution file op_rom_00015.bin.
libROM: Predicting solution at time 1.6000e+02 using ROM.
libROM: wallclock time: 0.241440 (seconds).
Writing solution file op_rom_00016.bin.
libROM: Predicting solution at time 1.7000e+02 using ROM.
libROM: wallclock time: 0.247398 (seconds).
Writing solution file op_rom_00017.bin.
libROM: Predicting solution at time 1.8000e+02 using ROM.
libROM: wallclock time: 0.244496 (seconds).
Writing solution file op_rom_00018.bin.
libROM: Predicting solution at time 1.9000e+02 using ROM.
libROM: wallclock time: 0.250613 (seconds).
Writing solution file op_rom_00019.bin.
libROM: Predicting solution at time 2.0000e+02 using ROM.
libROM: wallclock time: 0.246348 (seconds).
Writing solution file op_rom_00020.bin.
libROM: Predicting solution at time 2.1000e+02 using ROM.
libROM: wallclock time: 0.239400 (seconds).
Writing solution file op_rom_00021.bin.
libROM: Predicting solution at time 2.2000e+02 using ROM.
libROM: wallclock time: 0.239655 (seconds).
Writing solution file op_rom_00022.bin.
libROM: Predicting solution at time 2.3000e+02 using ROM.
libROM: wallclock time: 0.240208 (seconds).
Writing solution file op_rom_00023.bin.
libROM: Predicting solution at time 2.4000e+02 using ROM.
libROM: wallclock time: 0.243463 (seconds).
Writing solution file op_rom_00024.bin.
libROM: Predicting solution at time 2.5000e+02 using ROM.
libROM: wallclock time: 0.246869 (seconds).
Writing solution file op_rom_00025.bin.
libROM: Predicting solution at time 2.6000e+02 using ROM.
libROM: wallclock time: 0.238274 (seconds).
Writing solution file op_rom_00026.bin.
libROM: Predicting solution at time 2.7000e+02 using ROM.
libROM: wallclock time: 0.237310 (seconds).
Writing solution file op_rom_00027.bin.
libROM: Predicting solution at time 2.8000e+02 using ROM.
libROM: wallclock time: 0.236455 (seconds).
Writing solution file op_rom_00028.bin.
libROM: Predicting solution at time 2.9000e+02 using ROM.
libROM: wallclock time: 0.236396 (seconds).
Writing solution file op_rom_00029.bin.
libROM: Predicting solution at time 3.0000e+02 using ROM.
libROM: wallclock time: 0.239319 (seconds).
Writing solution file op_rom_00030.bin.
libROM: Predicting solution at time 3.1000e+02 using ROM.
libROM: wallclock time: 0.236808 (seconds).
Writing solution file op_rom_00031.bin.
libROM: Predicting solution at time 3.2000e+02 using ROM.
libROM: wallclock time: 0.238982 (seconds).
Writing solution file op_rom_00032.bin.
libROM: Predicting solution at time 3.3000e+02 using ROM.
libROM: wallclock time: 0.237195 (seconds).
Writing solution file op_rom_00033.bin.
libROM: Predicting solution at time 3.4000e+02 using ROM.
libROM: wallclock time: 0.238842 (seconds).
Writing solution file op_rom_00034.bin.
libROM: Predicting solution at time 3.5000e+02 using ROM.
libROM: wallclock time: 0.243906 (seconds).
Writing solution file op_rom_00035.bin.
libROM: Predicting solution at time 3.6000e+02 using ROM.
libROM: wallclock time: 0.241236 (seconds).
Writing solution file op_rom_00036.bin.
libROM: Predicting solution at time 3.7000e+02 using ROM.
libROM: wallclock time: 0.242927 (seconds).
Writing solution file op_rom_00037.bin.
libROM: Predicting solution at time 3.8000e+02 using ROM.
libROM: wallclock time: 0.236618 (seconds).
Writing solution file op_rom_00038.bin.
libROM: Predicting solution at time 3.9000e+02 using ROM.
libROM: wallclock time: 0.238566 (seconds).
Writing solution file op_rom_00039.bin.
libROM: Predicting solution at time 4.0000e+02 using ROM.
libROM: wallclock time: 0.238102 (seconds).
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom_00040.bin.
libROM: total prediction/query wallclock time: 9.836836 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object with filename root DMD/dmdobj_0000.
Saving DMD object with filename root DMD/dmdobj_0001.
Saving DMD object with filename root DMD/dmdobj_0002.
Saving DMD object with filename root DMD/dmdobj_0003.
Saving DMD object with filename root DMD/dmdobj_0004.
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 2.2563355954629010E-05
L2 Norm : 2.4733914130219971E-05
Linfinity Norm : 2.4975703864253711E-04
Solver runtime (in seconds): 4.5519722849999998E+03
Total runtime (in seconds): 4.5527609110000003E+03
Deallocating arrays.
Finished.