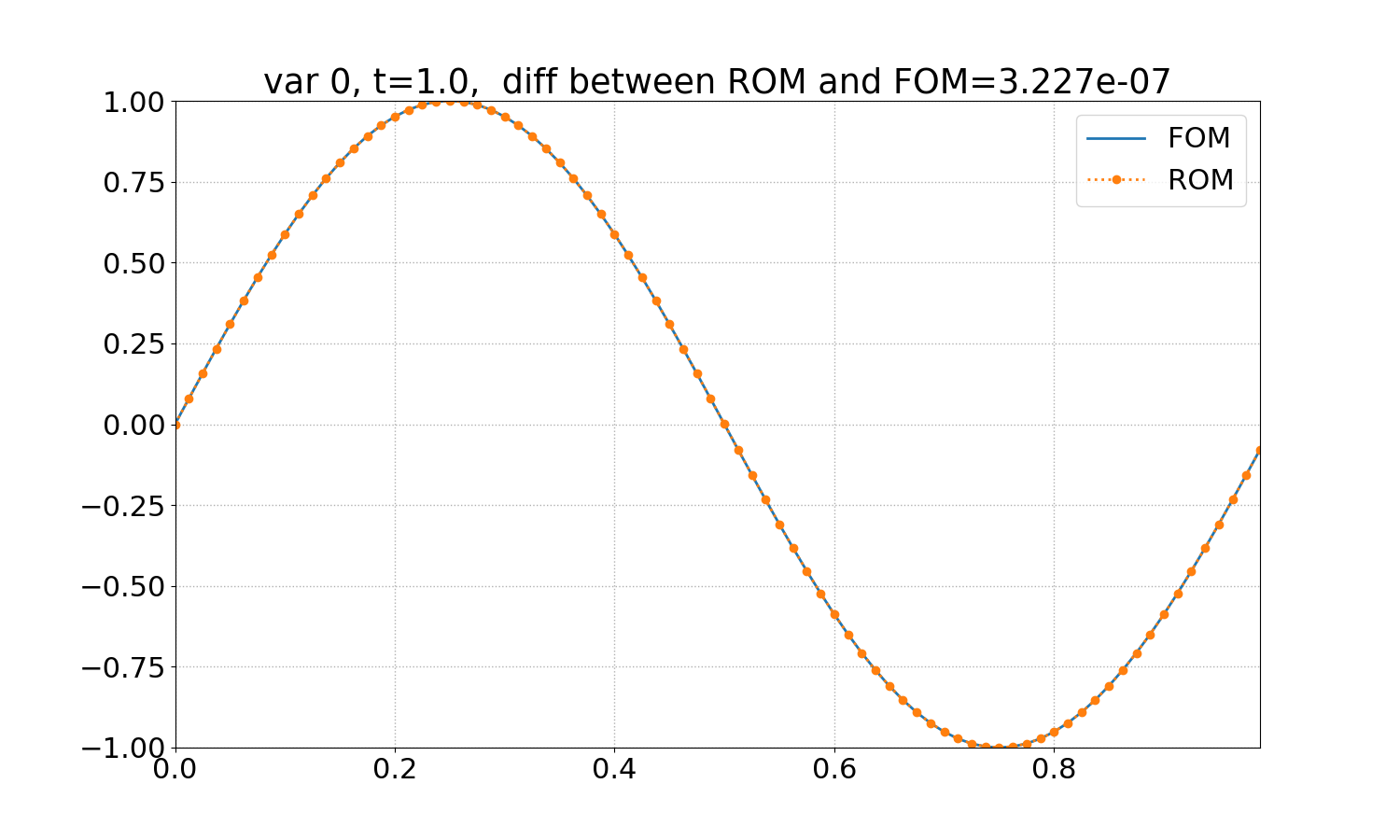
Reduced Order Modeling: This example trains a DMD object and then predicts the solution using the DMD at the same times that the actual HyPar solution is written at.
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
double pi = 4.0*atan(1.0);
int NI,ndims;
char ip_file_type[50];
strcpy(ip_file_type,"ascii");
FILE *in;
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) printf("Error: Input file \"solver.inp\" not found. Default values will be used.\n");
else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")){
while (strcmp(word, "end")){
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) fscanf(in,"%d",&ndims);
else if (!strcmp(word, "size")) fscanf(in,"%d",&NI);
else if (!strcmp(word, "ip_file_type")) fscanf(in,"%s",ip_file_type);
}
} else printf("Error: Illegal format in solver.inp. Crash and burn!\n");
}
fclose(in);
if (ndims != 1) {
printf("ndims is not 1 in solver.inp. this code is to generate 1D initial conditions\n");
return(0);
}
printf("Grid:\t\t\t%d\n",NI);
int i;
double dx = 1.0 / ((double)NI);
double *x, *u;
x = (double*) calloc (NI, sizeof(double));
u = (double*) calloc (NI, sizeof(double));
for (i = 0; i < NI; i++){
x[i] = i*dx;
u[i] = sin(2*pi*x[i]);
}
FILE *out;
if (!strcmp(ip_file_type,"ascii")) {
printf("Writing ASCII initial solution file initial.inp\n");
out = fopen("initial.inp","w");
for (i = 0; i < NI; i++) fprintf(out,"%lf ",x[i]);
fprintf(out,"\n");
for (i = 0; i < NI; i++) fprintf(out,"%lf ",u[i]);
fprintf(out,"\n");
fclose(out);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary initial solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x,sizeof(double),NI,out);
fwrite(u,sizeof(double),NI,out);
fclose(out);
}
free(x);
free(u);
return(0);
}
The first of each of these file sets is the solution at \(t=0\) and the final one is the solution at \(t=1\). Since HyPar::op_overwrite is set to no in solver.inp, a separate file is written for solutions at each output time. All the files are binary (HyPar::op_file_format is set to binary in solver.inp).
The following plot shows the final solution - FOM (full-order model) refers to the HyPar solution, ROM (reduced-order model) refers to the DMD solution.
By default, the code will write the trained DMD object(s) to files in a subdirectory (DMDROMObject::m_dirname - default value is "DMD"). If the subdirectory does not exist, the code may not report an error (or give some HDF5 file-writing error); the DMD objects will not be written! If the subdirectory exists, several files will exist after the simulation is complete - they are in a format that is readable by libROM.
HyPar - Parallel (MPI) version with 4 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 1
No. of variables : 1
Domain size : 80
Processes along each dimension : 4
Exact solution domain size : 80
No. of ghosts pts : 3
No. of iter. : 200
Restart iteration : 0
Time integration scheme : rk (ssprk3)
Spatial discretization scheme (hyperbolic) : crweno5
Split hyperbolic flux term? : no
Interpolation type for hyperbolic term : characteristic
Spatial discretization type (parabolic ) : nonconservative-1stage
Spatial discretization scheme (parabolic ) : 2
Time Step : 5.000000E-03
Check for conservation : yes
Screen output iterations : 10
File output iterations : 20
Initial solution file type : ascii
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : no
Physical model : linear-advection-diffusion-reaction
Partitioning domain and allocating data arrays.
Reading array from ASCII file initial.inp (Serial mode).
Volume integral of the initial solution:
0: -5.5511151231257827E-17
Reading boundary conditions from boundary.inp.
Boundary periodic: Along dimension 0 and face +1
Boundary periodic: Along dimension 0 and face -1
2 boundary condition(s) read.
Initializing solvers.
Reading WENO parameters from weno.inp.
Initializing physics. Model = "linear-advection-diffusion-reaction"
Reading physical model inputs from file "physics.inp".
Setting up time integration.
Setting up libROM interface.
libROM inputs and parameters:
reduced model dimensionality: 2
sampling frequency: 1
mode: train
type: DMD
save to file: true
local vector size: 20
libROM DMD inputs:
number of samples per window: 2147483647
directory name for DMD onjects: DMD
Solving in time (from 0 to 200 iterations)
Writing solution file op_00000.bin.
DMDROMObject::takeSample() - creating new DMD object, t=0.000000 (total: 1).
iter= 10 t=5.000E-02 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.2E-04 (s) cons_err=4.3368E-19
iter= 20 t=1.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.9E-04 (s) cons_err=4.3368E-19
Writing solution file op_00001.bin.
iter= 30 t=1.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.2E-04 (s) cons_err=2.5587E-17
iter= 40 t=2.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.2E-04 (s) cons_err=2.5587E-17
Writing solution file op_00002.bin.
iter= 50 t=2.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.6E-04 (s) cons_err=2.1684E-18
iter= 60 t=3.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.8E-04 (s) cons_err=6.5486E-17
Writing solution file op_00003.bin.
iter= 70 t=3.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.0E-04 (s) cons_err=8.1098E-17
iter= 80 t=4.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.1E-04 (s) cons_err=5.3343E-17
Writing solution file op_00004.bin.
iter= 90 t=4.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.1E-04 (s) cons_err=1.6176E-16
iter= 100 t=5.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.0E-04 (s) cons_err=6.0715E-17
Writing solution file op_00005.bin.
iter= 110 t=5.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.1E-04 (s) cons_err=4.8139E-17
iter= 120 t=6.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.8E-04 (s) cons_err=4.9006E-17
Writing solution file op_00006.bin.
iter= 130 t=6.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.0E-04 (s) cons_err=4.9006E-17
iter= 140 t=7.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.8E-04 (s) cons_err=1.0625E-16
Writing solution file op_00007.bin.
iter= 150 t=7.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.5E-04 (s) cons_err=7.6762E-17
iter= 160 t=8.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.0E-04 (s) cons_err=8.2399E-18
Writing solution file op_00008.bin.
iter= 170 t=8.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.7E-04 (s) cons_err=2.1250E-17
iter= 180 t=9.000E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.7E-04 (s) cons_err=4.7271E-17
Writing solution file op_00009.bin.
iter= 190 t=9.500E-01 CFL=4.000E-01 norm=2.2213E-02 wctime: 3.0E-04 (s) cons_err=4.5536E-17
iter= 200 t=1.000E+00 CFL=4.000E-01 norm=2.2213E-02 wctime: 2.8E-04 (s) cons_err=1.5699E-16
Completed time integration (Final time: 1.000000), total wctime: 0.718540 (seconds).
Reading array from ASCII file exact.inp (Serial mode).
Writing solution file op_00010.bin.
libROM: Training ROM.
DMDRomObject::train() - training DMD object 0 with 200 samples.
Using 2 basis vectors out of 80.
libROM: wallclock time: 0.059403 (seconds).
libROM: Predicting solution at time 0.0000e+00 using ROM.
libROM: wallclock time: 0.000077 (seconds).
Writing solution file op_rom_00000.bin.
libROM: Predicting solution at time 1.0000e-01 using ROM.
libROM: wallclock time: 0.000037 (seconds).
Writing solution file op_rom_00001.bin.
libROM: Predicting solution at time 2.0000e-01 using ROM.
libROM: wallclock time: 0.000030 (seconds).
Writing solution file op_rom_00002.bin.
libROM: Predicting solution at time 3.0000e-01 using ROM.
libROM: wallclock time: 0.000031 (seconds).
Writing solution file op_rom_00003.bin.
libROM: Predicting solution at time 4.0000e-01 using ROM.
libROM: wallclock time: 0.000033 (seconds).
Writing solution file op_rom_00004.bin.
libROM: Predicting solution at time 5.0000e-01 using ROM.
libROM: wallclock time: 0.000032 (seconds).
Writing solution file op_rom_00005.bin.
libROM: Predicting solution at time 6.0000e-01 using ROM.
libROM: wallclock time: 0.000030 (seconds).
Writing solution file op_rom_00006.bin.
libROM: Predicting solution at time 7.0000e-01 using ROM.
libROM: wallclock time: 0.000047 (seconds).
Writing solution file op_rom_00007.bin.
libROM: Predicting solution at time 8.0000e-01 using ROM.
libROM: wallclock time: 0.000030 (seconds).
Writing solution file op_rom_00008.bin.
libROM: Predicting solution at time 9.0000e-01 using ROM.
libROM: wallclock time: 0.000030 (seconds).
Writing solution file op_rom_00009.bin.
libROM: Predicting solution at time 1.0000e+00 using ROM.
libROM: wallclock time: 0.000029 (seconds).
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom_00010.bin.
libROM: total prediction/query wallclock time: 0.000406 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object with filename root DMD/dmdobj_0000.
Computed errors for domain 0:
L1 Error : 8.1564188969988377E-06
L2 Error : 8.1563255924379997E-06
Linfinity Error : 8.4640413008374438E-06
Conservation Errors:
1.5699247457590104E-16
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 2.9065523787146478E-07
L2 Norm : 3.2037250962276641E-07
Linfinity Norm : 4.1231337540346448E-07
Solver runtime (in seconds): 2.4540210000000000E+00
Total runtime (in seconds): 3.2191299999999998E+00
Deallocating arrays.
Finished.