Initial solution: A warm bubble in cool ambient atmosphere. Note that in this example, the gravitational forces and rising of the bubble is along the y-axis.
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
#include <string.h>
{
return(exp(a*log(x)));
}
{
double gamma = 1.4;
double R = 287.058;
double rho_ref = 1.1612055171196529;
double p_ref = 100000.0;
double grav_x = 0.0;
double grav_y = 9.8;
double grav_z = 0.0;
int HB = 0;
int NI,NJ,NK,ndims;
char ip_file_type[50]; strcpy(ip_file_type,"ascii");
FILE *in;
printf("Reading file \"solver.inp\"...\n");
in = fopen("solver.inp","r");
if (!in) {
printf("Error: Input file \"solver.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")) {
while (strcmp(word, "end")) {
fscanf(in,"%s",word);
if (!strcmp(word, "ndims")) fscanf(in,"%d",&ndims);
else if (!strcmp(word, "size")) {
fscanf(in,"%d",&NI);
fscanf(in,"%d",&NJ);
fscanf(in,"%d",&NK);
} else if (!strcmp(word, "ip_file_type")) fscanf(in,"%s",ip_file_type);
}
} else printf("Error: Illegal format in solver.inp. Crash and burn!\n");
}
fclose(in);
printf("Reading file \"physics.inp\"...\n");
in = fopen("physics.inp","r");
if (!in) {
printf("Error: Input file \"physics.inp\" not found.\n");
return(0);
} else {
char word[500];
fscanf(in,"%s",word);
if (!strcmp(word, "begin")) {
while (strcmp(word, "end")) {
fscanf(in,"%s",word);
if (!strcmp(word, "rho_ref")) fscanf(in,"%lf",&rho_ref);
else if (!strcmp(word, "p_ref" )) fscanf(in,"%lf",&p_ref );
else if (!strcmp(word, "gamma" )) fscanf(in,"%lf",&gamma );
else if (!strcmp(word, "R" )) fscanf(in,"%lf",&R );
else if (!strcmp(word, "HB" )) fscanf(in,"%d" ,&HB );
else if (!strcmp(word, "gravity")) {
fscanf(in,"%lf",&grav_x );
fscanf(in,"%lf",&grav_y );
fscanf(in,"%lf",&grav_z );
}
}
} else printf("Error: Illegal format in physics.inp. Crash and burn!\n");
}
fclose(in);
if (ndims != 3) {
printf("ndims is not 3 in solver.inp. this code is to generate 3D initial conditions\n");
return(0);
}
if (HB != 2) {
printf("Error: Specify \"HB\" as 2 in physics.inp.\n");
}
if ((grav_x != 0.0) || (grav_z != 0.0)) {
printf("Error: gravity must be zero along x and z for this example.\n");
return(0);
}
printf("Grid:\t\t\t%d X %d X %d\n",NI,NJ,NK);
printf("Reference density and pressure: %lf, %lf.\n",rho_ref,p_ref);
int i,j,k;
double dx = 1000.0 / ((double)(NI-1));
double dy = 1000.0 / ((double)(NJ-1));
double dz = 1000.0 / ((double)(NK-1));
double *x, *y, *z, *U;
x = (double*) calloc (NI , sizeof(double));
y = (double*) calloc (NJ , sizeof(double));
z = (double*) calloc (NK , sizeof(double));
U = (double*) calloc (5*NI*NJ*NK, sizeof(double));
double xc = 500;
double yc = 260;
double zc = 500;
double Cp = gamma * R / (gamma-1.0);
double pi = 4.0*atan(1.0);
double rc = 250.0;
double T_ref = p_ref / (R * rho_ref);
for (i = 0; i < NI; i++){
for (j = 0; j < NJ; j++){
for (k = 0; k < NK; k++){
x[i] = i*dx;
y[j] = j*dy;
z[k] = k*dz;
int p = i + NI*j + NI*NJ*k;
double r = sqrt((x[i]-xc)*(x[i]-xc)+(y[j]-yc)*(y[j]-yc)+(z[k]-zc)*(z[k]-zc));
double dtheta = (r>rc ? 0.0 : (0.5*(1.0+cos(pi*r/rc))) );
double theta = T_ref + dtheta;
double Pexner = 1.0 - (grav_y*y[j])/(Cp*T_ref);
double rho = (p_ref/(R*theta)) *
raiseto(Pexner, (1.0/(gamma-1.0)));
double E = rho * (R/(gamma-1.0)) * theta*Pexner;
U[5*p+0] = rho;
U[5*p+1] = 0.0;
U[5*p+2] = 0.0;
U[5*p+3] = 0.0;
U[5*p+4] = E;
}
}
}
FILE *out;
if (!strcmp(ip_file_type,"ascii")) {
printf("Error: sorry, ASCII format initial solution file not implemented. ");
printf("Please choose \"ip_file_format\" in solver.inp as \"binary\".\n");
return(0);
} else if ((!strcmp(ip_file_type,"binary")) || (!strcmp(ip_file_type,"bin"))) {
printf("Writing binary initial solution file initial.inp\n");
out = fopen("initial.inp","wb");
fwrite(x,sizeof(double),NI,out);
fwrite(y,sizeof(double),NJ,out);
fwrite(z,sizeof(double),NK,out);
fwrite(U,sizeof(double),5*NI*NJ*NK,out);
fclose(out);
}
free(x);
free(y);
free(z);
free(U);
return(0);
}
The binary solution file contains the conserved variables ( \(\rho,\rho u,\rho v,\rho w,e\)).
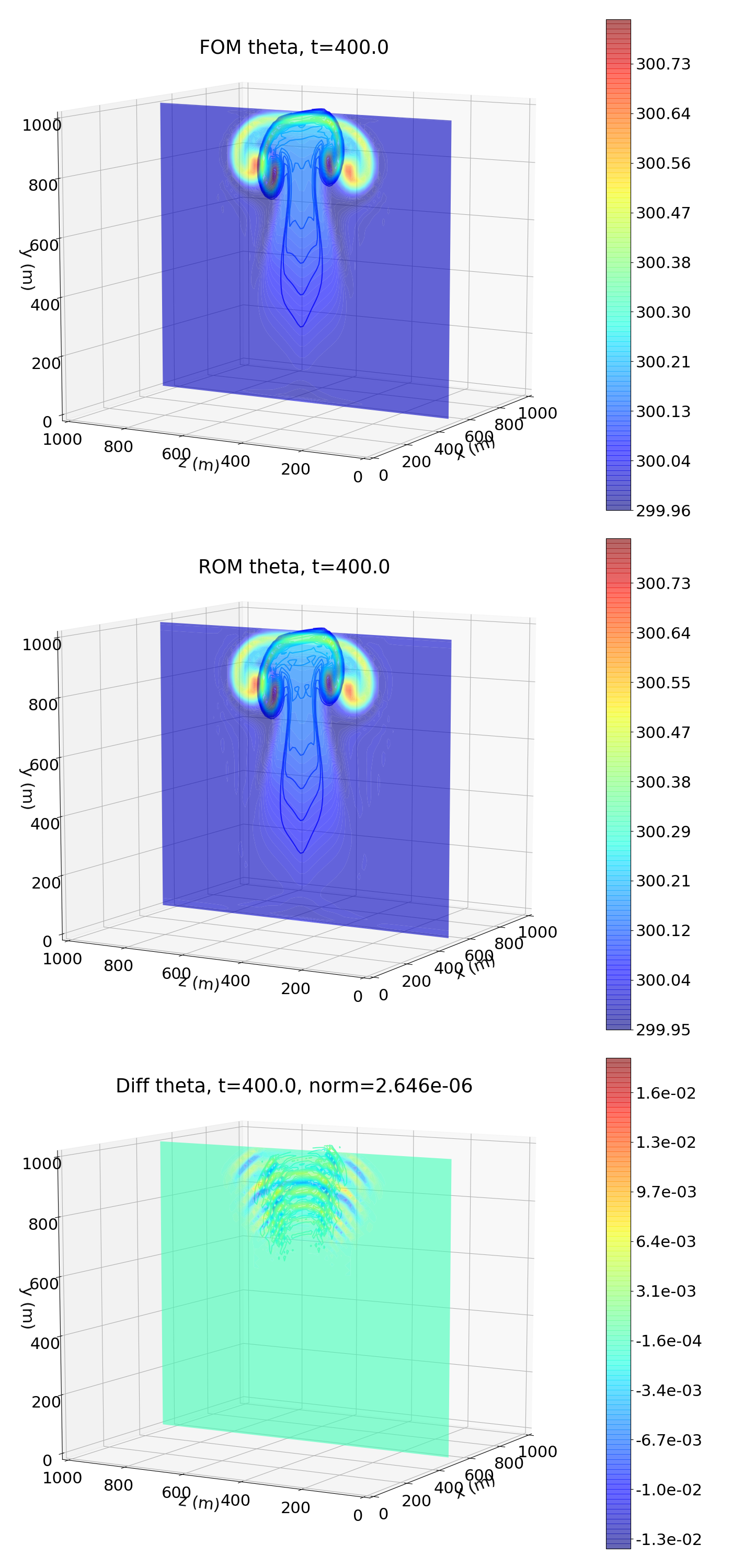
The following figure shows the potential temperature at the final time. Colored contours are plotted in the X-Y plane, and contour lines are plotted in the Z-Y plane (FOM is the HyPar solution, ROM is the DMD prediction, and diff is the difference between the two):
srun: job 10240594 queued and waiting for resources
srun: job 10240594 has been allocated resources
HyPar - Parallel (MPI) version with 64 processes
Compiled with PETSc time integration.
Allocated simulation object(s).
Reading solver inputs from file "solver.inp".
No. of dimensions : 3
No. of variables : 5
Domain size : 64 64 64
Processes along each dimension : 4 4 4
Exact solution domain size : 64 64 64
No. of ghosts pts : 3
No. of iter. : 200
Restart iteration : 0
Time integration scheme : PETSc
Spatial discretization scheme (hyperbolic) : weno5
Split hyperbolic flux term? : yes
Interpolation type for hyperbolic term : components
Spatial discretization type (parabolic ) : nonconservative-1stage
Spatial discretization scheme (parabolic ) : 2
Time Step : 2.000000E+00
Check for conservation : no
Screen output iterations : 2
File output iterations : 20
Initial solution file type : binary
Initial solution read mode : serial
Solution file write mode : serial
Solution file format : binary
Overwrite solution file : no
Physical model : navierstokes3d
Partitioning domain and allocating data arrays.
Reading array from binary file initial.inp (Serial mode).
Volume integral of the initial solution:
0: 1.1686650873135223E+09
1: 0.0000000000000000E+00
2: 0.0000000000000000E+00
3: 0.0000000000000000E+00
4: 2.4758406164981962E+14
Reading boundary conditions from boundary.inp.
Boundary slip-wall: Along dimension 0 and face +1
Boundary slip-wall: Along dimension 0 and face -1
Boundary slip-wall: Along dimension 1 and face +1
Boundary slip-wall: Along dimension 1 and face -1
Boundary slip-wall: Along dimension 2 and face +1
Boundary slip-wall: Along dimension 2 and face -1
6 boundary condition(s) read.
Initializing solvers.
Reading WENO parameters from weno.inp.
Initializing physics. Model = "navierstokes3d"
Reading physical model inputs from file "physics.inp".
Setting up PETSc time integration...
Setting up libROM interface.
libROMInterface inputs and parameters:
reduced model dimensionality: 16
sampling frequency: 1
mode: train
component mode: monolithic
type: DMD
save to file: true
DMDROMObject details:
number of samples per window: 50
directory name for DMD onjects: DMD
write snapshot matrix to file: false
simulation domain: 0
PETSc: total number of computational points is 262144.
PETSc: total number of computational DOFs is 1310720.
Implicit-Explicit time-integration:-
Hyperbolic (f-df) term: Explicit
Hyperbolic (df) term: Implicit
Parabolic term: Implicit
Source term: Implicit
SolvePETSc(): Problem type is linear.
** Starting PETSc time integration **
Writing solution file op_00000.bin.
DMDROMObject::takeSample() - creating new DMD object for sim. domain 0, var -1, t=0.000000 (total: 1).
iter= 2 dt=2.000E+00 t=4.000E+00 CFL=4.375E+01 norm=4.6613E-01 wctime: 3.7E+00 (s)
iter= 4 dt=2.000E+00 t=8.000E+00 CFL=4.375E+01 norm=1.7872E-01 wctime: 3.8E+00 (s)
iter= 6 dt=2.000E+00 t=1.200E+01 CFL=4.375E+01 norm=3.0277E-01 wctime: 3.8E+00 (s)
iter= 8 dt=2.000E+00 t=1.600E+01 CFL=4.376E+01 norm=4.0854E-01 wctime: 4.0E+00 (s)
iter= 10 dt=2.000E+00 t=2.000E+01 CFL=4.376E+01 norm=3.8615E-01 wctime: 3.8E+00 (s)
iter= 12 dt=2.000E+00 t=2.400E+01 CFL=4.376E+01 norm=2.7133E-01 wctime: 3.7E+00 (s)
iter= 14 dt=2.000E+00 t=2.800E+01 CFL=4.376E+01 norm=9.6926E-02 wctime: 4.0E+00 (s)
iter= 16 dt=2.000E+00 t=3.200E+01 CFL=4.376E+01 norm=7.2168E-02 wctime: 4.0E+00 (s)
iter= 18 dt=2.000E+00 t=3.600E+01 CFL=4.376E+01 norm=2.0010E-01 wctime: 4.2E+00 (s)
iter= 20 dt=2.000E+00 t=4.000E+01 CFL=4.376E+01 norm=2.5134E-01 wctime: 4.1E+00 (s)
Writing solution file op_00001.bin.
iter= 22 dt=2.000E+00 t=4.400E+01 CFL=4.377E+01 norm=2.3351E-01 wctime: 3.8E+00 (s)
iter= 24 dt=2.000E+00 t=4.800E+01 CFL=4.377E+01 norm=1.5111E-01 wctime: 3.8E+00 (s)
iter= 26 dt=2.000E+00 t=5.200E+01 CFL=4.378E+01 norm=4.7897E-02 wctime: 4.1E+00 (s)
iter= 28 dt=2.000E+00 t=5.600E+01 CFL=4.379E+01 norm=6.0644E-02 wctime: 4.1E+00 (s)
iter= 30 dt=2.000E+00 t=6.000E+01 CFL=4.379E+01 norm=1.2829E-01 wctime: 4.5E+00 (s)
iter= 32 dt=2.000E+00 t=6.400E+01 CFL=4.380E+01 norm=1.5991E-01 wctime: 4.1E+00 (s)
iter= 34 dt=2.000E+00 t=6.800E+01 CFL=4.381E+01 norm=1.3642E-01 wctime: 4.0E+00 (s)
iter= 36 dt=2.000E+00 t=7.200E+01 CFL=4.381E+01 norm=8.9307E-02 wctime: 4.0E+00 (s)
iter= 38 dt=2.000E+00 t=7.600E+01 CFL=4.382E+01 norm=1.9225E-02 wctime: 4.3E+00 (s)
iter= 40 dt=2.000E+00 t=8.000E+01 CFL=4.383E+01 norm=4.2010E-02 wctime: 4.3E+00 (s)
Writing solution file op_00002.bin.
iter= 42 dt=2.000E+00 t=8.400E+01 CFL=4.383E+01 norm=8.8311E-02 wctime: 4.4E+00 (s)
iter= 44 dt=2.000E+00 t=8.800E+01 CFL=4.384E+01 norm=9.5145E-02 wctime: 4.3E+00 (s)
iter= 46 dt=2.000E+00 t=9.200E+01 CFL=4.384E+01 norm=8.6380E-02 wctime: 4.2E+00 (s)
iter= 48 dt=2.000E+00 t=9.600E+01 CFL=4.385E+01 norm=4.6742E-02 wctime: 4.2E+00 (s)
iter= 50 dt=2.000E+00 t=1.000E+02 CFL=4.385E+01 norm=1.6582E-02 wctime: 4.4E+00 (s)
DMDROMObject::train() - training DMD object 0 for sim. domain 0, var -1 with 51 samples.
Using 16 basis vectors out of 50.
DMDROMObject::takeSample() - creating new DMD object for sim. domain 0, var -1, t=100.000000 (total: 2).
iter= 52 dt=2.000E+00 t=1.040E+02 CFL=4.386E+01 norm=3.7296E-02 wctime: 4.4E+00 (s)
iter= 54 dt=2.000E+00 t=1.080E+02 CFL=4.386E+01 norm=5.3518E-02 wctime: 4.5E+00 (s)
iter= 56 dt=2.000E+00 t=1.120E+02 CFL=4.387E+01 norm=6.5068E-02 wctime: 4.4E+00 (s)
iter= 58 dt=2.000E+00 t=1.160E+02 CFL=4.387E+01 norm=4.8353E-02 wctime: 4.4E+00 (s)
iter= 60 dt=2.000E+00 t=1.200E+02 CFL=4.388E+01 norm=3.4015E-02 wctime: 4.4E+00 (s)
Writing solution file op_00003.bin.
iter= 62 dt=2.000E+00 t=1.240E+02 CFL=4.388E+01 norm=1.8027E-02 wctime: 4.6E+00 (s)
iter= 64 dt=2.000E+00 t=1.280E+02 CFL=4.388E+01 norm=2.6413E-02 wctime: 4.6E+00 (s)
iter= 66 dt=2.000E+00 t=1.320E+02 CFL=4.388E+01 norm=4.2508E-02 wctime: 4.5E+00 (s)
iter= 68 dt=2.000E+00 t=1.360E+02 CFL=4.389E+01 norm=3.9334E-02 wctime: 4.6E+00 (s)
iter= 70 dt=2.000E+00 t=1.400E+02 CFL=4.389E+01 norm=3.7670E-02 wctime: 4.7E+00 (s)
iter= 72 dt=2.000E+00 t=1.440E+02 CFL=4.389E+01 norm=2.4546E-02 wctime: 4.6E+00 (s)
iter= 74 dt=2.000E+00 t=1.480E+02 CFL=4.389E+01 norm=2.2579E-02 wctime: 4.6E+00 (s)
iter= 76 dt=2.000E+00 t=1.520E+02 CFL=4.389E+01 norm=2.9157E-02 wctime: 4.6E+00 (s)
iter= 78 dt=2.000E+00 t=1.560E+02 CFL=4.389E+01 norm=3.1504E-02 wctime: 4.6E+00 (s)
iter= 80 dt=2.000E+00 t=1.600E+02 CFL=4.389E+01 norm=3.5025E-02 wctime: 4.6E+00 (s)
Writing solution file op_00004.bin.
iter= 82 dt=2.000E+00 t=1.640E+02 CFL=4.390E+01 norm=3.0110E-02 wctime: 4.6E+00 (s)
iter= 84 dt=2.000E+00 t=1.680E+02 CFL=4.390E+01 norm=2.8470E-02 wctime: 4.6E+00 (s)
iter= 86 dt=2.000E+00 t=1.720E+02 CFL=4.390E+01 norm=2.8242E-02 wctime: 4.5E+00 (s)
iter= 88 dt=2.000E+00 t=1.760E+02 CFL=4.390E+01 norm=3.0403E-02 wctime: 4.6E+00 (s)
iter= 90 dt=2.000E+00 t=1.800E+02 CFL=4.389E+01 norm=3.3249E-02 wctime: 4.6E+00 (s)
iter= 92 dt=2.000E+00 t=1.840E+02 CFL=4.389E+01 norm=3.3613E-02 wctime: 4.5E+00 (s)
iter= 94 dt=2.000E+00 t=1.880E+02 CFL=4.389E+01 norm=3.3177E-02 wctime: 4.5E+00 (s)
iter= 96 dt=2.000E+00 t=1.920E+02 CFL=4.389E+01 norm=3.3013E-02 wctime: 4.6E+00 (s)
iter= 98 dt=2.000E+00 t=1.960E+02 CFL=4.389E+01 norm=3.3826E-02 wctime: 4.5E+00 (s)
iter= 100 dt=2.000E+00 t=2.000E+02 CFL=4.389E+01 norm=3.5022E-02 wctime: 4.5E+00 (s)
Writing solution file op_00005.bin.
DMDROMObject::train() - training DMD object 1 for sim. domain 0, var -1 with 51 samples.
Using 16 basis vectors out of 50.
DMDROMObject::takeSample() - creating new DMD object for sim. domain 0, var -1, t=200.000000 (total: 3).
iter= 102 dt=2.000E+00 t=2.040E+02 CFL=4.389E+01 norm=3.7046E-02 wctime: 4.5E+00 (s)
iter= 104 dt=2.000E+00 t=2.080E+02 CFL=4.389E+01 norm=3.6900E-02 wctime: 4.5E+00 (s)
iter= 106 dt=2.000E+00 t=2.120E+02 CFL=4.389E+01 norm=3.8191E-02 wctime: 4.5E+00 (s)
iter= 108 dt=2.000E+00 t=2.160E+02 CFL=4.389E+01 norm=3.8205E-02 wctime: 4.6E+00 (s)
iter= 110 dt=2.000E+00 t=2.200E+02 CFL=4.388E+01 norm=3.9083E-02 wctime: 4.5E+00 (s)
iter= 112 dt=2.000E+00 t=2.240E+02 CFL=4.388E+01 norm=4.0808E-02 wctime: 4.5E+00 (s)
iter= 114 dt=2.000E+00 t=2.280E+02 CFL=4.388E+01 norm=4.0738E-02 wctime: 4.5E+00 (s)
iter= 116 dt=2.000E+00 t=2.320E+02 CFL=4.388E+01 norm=4.2765E-02 wctime: 4.5E+00 (s)
iter= 118 dt=2.000E+00 t=2.360E+02 CFL=4.388E+01 norm=4.2462E-02 wctime: 4.5E+00 (s)
iter= 120 dt=2.000E+00 t=2.400E+02 CFL=4.388E+01 norm=4.3767E-02 wctime: 4.5E+00 (s)
Writing solution file op_00006.bin.
iter= 122 dt=2.000E+00 t=2.440E+02 CFL=4.387E+01 norm=4.4773E-02 wctime: 4.5E+00 (s)
iter= 124 dt=2.000E+00 t=2.480E+02 CFL=4.387E+01 norm=4.5055E-02 wctime: 4.5E+00 (s)
iter= 126 dt=2.000E+00 t=2.520E+02 CFL=4.387E+01 norm=4.7022E-02 wctime: 4.5E+00 (s)
iter= 128 dt=2.000E+00 t=2.560E+02 CFL=4.387E+01 norm=4.6817E-02 wctime: 4.6E+00 (s)
iter= 130 dt=2.000E+00 t=2.600E+02 CFL=4.387E+01 norm=4.8519E-02 wctime: 4.6E+00 (s)
iter= 132 dt=2.000E+00 t=2.640E+02 CFL=4.387E+01 norm=4.9059E-02 wctime: 4.6E+00 (s)
iter= 134 dt=2.000E+00 t=2.680E+02 CFL=4.386E+01 norm=4.9884E-02 wctime: 4.6E+00 (s)
iter= 136 dt=2.000E+00 t=2.720E+02 CFL=4.386E+01 norm=5.1468E-02 wctime: 4.6E+00 (s)
iter= 138 dt=2.000E+00 t=2.760E+02 CFL=4.386E+01 norm=5.1688E-02 wctime: 4.6E+00 (s)
iter= 140 dt=2.000E+00 t=2.800E+02 CFL=4.386E+01 norm=5.3510E-02 wctime: 4.6E+00 (s)
Writing solution file op_00007.bin.
iter= 142 dt=2.000E+00 t=2.840E+02 CFL=4.386E+01 norm=5.4022E-02 wctime: 4.6E+00 (s)
iter= 144 dt=2.000E+00 t=2.880E+02 CFL=4.386E+01 norm=5.5372E-02 wctime: 4.6E+00 (s)
iter= 146 dt=2.000E+00 t=2.920E+02 CFL=4.386E+01 norm=5.6722E-02 wctime: 4.6E+00 (s)
iter= 148 dt=2.000E+00 t=2.960E+02 CFL=4.385E+01 norm=5.7594E-02 wctime: 4.7E+00 (s)
iter= 150 dt=2.000E+00 t=3.000E+02 CFL=4.385E+01 norm=5.9456E-02 wctime: 4.6E+00 (s)
DMDROMObject::train() - training DMD object 2 for sim. domain 0, var -1 with 51 samples.
Using 16 basis vectors out of 50.
DMDROMObject::takeSample() - creating new DMD object for sim. domain 0, var -1, t=300.000000 (total: 4).
iter= 152 dt=2.000E+00 t=3.040E+02 CFL=4.385E+01 norm=6.0386E-02 wctime: 4.6E+00 (s)
iter= 154 dt=2.000E+00 t=3.080E+02 CFL=4.385E+01 norm=6.2173E-02 wctime: 4.6E+00 (s)
iter= 156 dt=2.000E+00 t=3.120E+02 CFL=4.385E+01 norm=6.3620E-02 wctime: 4.6E+00 (s)
iter= 158 dt=2.000E+00 t=3.160E+02 CFL=4.385E+01 norm=6.5114E-02 wctime: 4.6E+00 (s)
iter= 160 dt=2.000E+00 t=3.200E+02 CFL=4.385E+01 norm=6.7011E-02 wctime: 4.6E+00 (s)
Writing solution file op_00008.bin.
iter= 162 dt=2.000E+00 t=3.240E+02 CFL=4.385E+01 norm=6.8407E-02 wctime: 4.6E+00 (s)
iter= 164 dt=2.000E+00 t=3.280E+02 CFL=4.384E+01 norm=7.0360E-02 wctime: 4.6E+00 (s)
iter= 166 dt=2.000E+00 t=3.320E+02 CFL=4.384E+01 norm=7.1921E-02 wctime: 4.6E+00 (s)
iter= 168 dt=2.000E+00 t=3.360E+02 CFL=4.384E+01 norm=7.3650E-02 wctime: 4.6E+00 (s)
iter= 170 dt=2.000E+00 t=3.400E+02 CFL=4.384E+01 norm=7.5378E-02 wctime: 4.6E+00 (s)
iter= 172 dt=2.000E+00 t=3.440E+02 CFL=4.384E+01 norm=7.6843E-02 wctime: 4.7E+00 (s)
iter= 174 dt=2.000E+00 t=3.480E+02 CFL=4.384E+01 norm=7.8483E-02 wctime: 4.6E+00 (s)
iter= 176 dt=2.000E+00 t=3.520E+02 CFL=4.384E+01 norm=7.9745E-02 wctime: 4.6E+00 (s)
iter= 178 dt=2.000E+00 t=3.560E+02 CFL=4.384E+01 norm=8.1031E-02 wctime: 4.6E+00 (s)
iter= 180 dt=2.000E+00 t=3.600E+02 CFL=4.383E+01 norm=8.2083E-02 wctime: 4.6E+00 (s)
Writing solution file op_00009.bin.
iter= 182 dt=2.000E+00 t=3.640E+02 CFL=4.383E+01 norm=8.2910E-02 wctime: 4.6E+00 (s)
iter= 184 dt=2.000E+00 t=3.680E+02 CFL=4.383E+01 norm=8.3637E-02 wctime: 4.6E+00 (s)
iter= 186 dt=2.000E+00 t=3.720E+02 CFL=4.383E+01 norm=8.4031E-02 wctime: 4.6E+00 (s)
iter= 188 dt=2.000E+00 t=3.760E+02 CFL=4.382E+01 norm=8.4328E-02 wctime: 4.6E+00 (s)
iter= 190 dt=2.000E+00 t=3.800E+02 CFL=4.382E+01 norm=8.4360E-02 wctime: 4.6E+00 (s)
iter= 192 dt=2.000E+00 t=3.840E+02 CFL=4.381E+01 norm=8.4218E-02 wctime: 4.6E+00 (s)
iter= 194 dt=2.000E+00 t=3.880E+02 CFL=4.381E+01 norm=8.3925E-02 wctime: 4.6E+00 (s)
iter= 196 dt=2.000E+00 t=3.920E+02 CFL=4.380E+01 norm=8.3438E-02 wctime: 4.7E+00 (s)
iter= 198 dt=2.000E+00 t=3.960E+02 CFL=4.380E+01 norm=8.2870E-02 wctime: 4.7E+00 (s)
iter= 200 dt=2.000E+00 t=4.000E+02 CFL=4.379E+01 norm=8.2161E-02 wctime: 4.7E+00 (s)
Writing solution file op_00010.bin.
** Completed PETSc time integration (Final time: 400.000000), total wctime: 906.731281 (seconds) **
DMDROMObject::train() - training DMD object 3 for sim. domain 0, var -1 with 50 samples.
Using 16 basis vectors out of 49.
libROM: total training wallclock time: 22.486292 (seconds).
libROM: Predicted solution at time 0.0000e+00 using ROM, wallclock time: 0.273265.
Writing solution file op_rom_00000.bin.
libROM: Predicted solution at time 4.0000e+01 using ROM, wallclock time: 0.410028.
Writing solution file op_rom_00001.bin.
libROM: Predicted solution at time 8.0000e+01 using ROM, wallclock time: 0.485338.
Writing solution file op_rom_00002.bin.
libROM: Predicted solution at time 1.2000e+02 using ROM, wallclock time: 0.379907.
Writing solution file op_rom_00003.bin.
libROM: Predicted solution at time 1.6000e+02 using ROM, wallclock time: 0.398802.
Writing solution file op_rom_00004.bin.
libROM: Predicted solution at time 2.0000e+02 using ROM, wallclock time: 0.383941.
Writing solution file op_rom_00005.bin.
libROM: Predicted solution at time 2.4000e+02 using ROM, wallclock time: 0.376884.
Writing solution file op_rom_00006.bin.
libROM: Predicted solution at time 2.8000e+02 using ROM, wallclock time: 0.383239.
Writing solution file op_rom_00007.bin.
libROM: Predicted solution at time 3.2000e+02 using ROM, wallclock time: 0.365753.
Writing solution file op_rom_00008.bin.
libROM: Predicted solution at time 3.6000e+02 using ROM, wallclock time: 0.365955.
Writing solution file op_rom_00009.bin.
libROM: Predicted solution at time 4.0000e+02 using ROM, wallclock time: 0.365799.
Writing solution file op_rom_00010.bin.
libROM: Predicted solution at time 4.0000e+02 using ROM, wallclock time: 0.365992.
libROM: Calculating diff between PDE and ROM solutions.
Writing solution file op_rom_00011.bin.
libROM: total prediction/query wallclock time: 4.554903 (seconds).
libROMInterface::saveROM() - saving ROM objects.
Saving DMD object and summary (DMD/dmdobj_0000, DMD/dmd_summary_0000).
Saving DMD object and summary (DMD/dmdobj_0001, DMD/dmd_summary_0001).
Saving DMD object and summary (DMD/dmdobj_0002, DMD/dmd_summary_0002).
Saving DMD object and summary (DMD/dmdobj_0003, DMD/dmd_summary_0003).
Norms of the diff between ROM and PDE solutions for domain 0:
L1 Norm : 3.5061348189940579E-09
L2 Norm : 8.1771689835182761E-09
Linfinity Norm : 1.8798434168943727E-07
Solver runtime (in seconds): 9.3870696399999997E+02
Total runtime (in seconds): 9.3893929700000001E+02
Deallocating arrays.
Finished.